6FBQ
 
 | | Crystal Structure of the Human Retinoid X Receptor DNA-Binding Domain Bound to the Human MEp DR1 Response Element, pH 7.0 | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, DNA (5'-D(*CP*TP*GP*GP*GP*TP*CP*AP*AP*AP*GP*TP*TP*CP*AP*TP*C)-3'), DNA (5'-D(*GP*AP*TP*GP*AP*AP*CP*TP*TP*TP*GP*AP*CP*CP*CP*AP*G)-3'), ... | | Authors: | McEwen, A.G, Poussin-Courmontagne, P, Osz, J, Rochel, N. | | Deposit date: | 2017-12-19 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Modulation of RXR-DNA complex assembly by DNA context.
Mol. Cell. Endocrinol., 481, 2019
|
|
6FBR
 
 | | Crystal Structure of the Human Retinoid X Receptor DNA-Binding Domain Bound to the Human MEp DR1 Response Element, pH 4.2 | | Descriptor: | DI(HYDROXYETHYL)ETHER, DNA (5'-D(*CP*TP*GP*GP*GP*TP*CP*AP*AP*AP*GP*TP*TP*CP*AP*TP*C)-3'), DNA (5'-D(*GP*AP*TP*GP*AP*AP*CP*TP*TP*TP*GP*AP*CP*CP*CP*AP*G)-3'), ... | | Authors: | McEwen, A.G, Poussin-Courmontagne, P, Osz, J, Rochel, N. | | Deposit date: | 2017-12-19 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Modulation of RXR-DNA complex assembly by DNA context.
Mol. Cell. Endocrinol., 481, 2019
|
|
4GIZ
 
 | | Crystal structure of full-length human papillomavirus oncoprotein E6 in complex with LXXLL peptide of ubiquitin ligase E6AP at 2.55 A resolution | | Descriptor: | Maltose-binding periplasmic protein, UBIQUITIN LIGASE EA6P: chimeric protein, Protein E6, ... | | Authors: | McEwen, A.G, Zanier, K, Charbonnier, S, Poussin, P, Cura, V, Vande Pol, S, Trave, G, Cavarelli, J. | | Deposit date: | 2012-08-09 | | Release date: | 2013-01-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural basis for hijacking of cellular LxxLL motifs by papillomavirus E6 oncoproteins.
Science, 339, 2013
|
|
6FC9
 
 | | The 1,8-bis(aminomethyl)anthracene and Quadruplex-duplex junction complex | | Descriptor: | DNA (27-MER), [8-(azaniumylmethyl)anthracen-1-yl]methylazanium | | Authors: | Santana, A, Serrano, I, Montalvillo-Jimenez, L, Corzana, F, Bastida, A, Jimenez-Barbero, J, Gonzalez, C, Asensio, J.L. | | Deposit date: | 2017-12-20 | | Release date: | 2019-04-10 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | De Novo Design of Selective Quadruplex-Duplex Junction Ligands and Structural Characterisation of Their Binding Mode: Targeting the G4 Hot-Spot.
Chemistry, 2020
|
|
6FOI
 
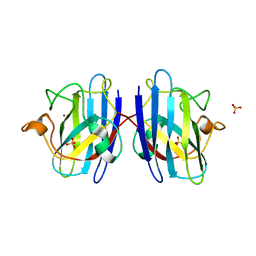 | | Human Cys57/156Ala superoxide dismutase-1 (SOD1), as isolated. | | Descriptor: | SULFATE ION, Superoxide dismutase [Cu-Zn], ZINC ION | | Authors: | Wright, G.S.A, Sala, F.A, Antonyuk, S.V, Garratt, R.C, Hasnain, S.S. | | Deposit date: | 2018-02-07 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular recognition and maturation of SOD1 by its evolutionarily destabilised cognate chaperone hCCS.
Plos Biol., 17, 2019
|
|
6FON
 
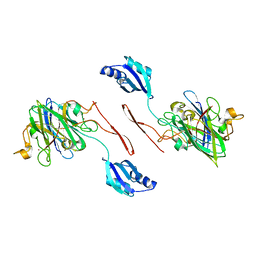 | | Elongated conformer of the human copper chaperone for SOD1 complexed with human SOD1 | | Descriptor: | Copper chaperone for superoxide dismutase, Superoxide dismutase [Cu-Zn], ZINC ION | | Authors: | Wright, G.S.A, Sala, F.A, Antonyuk, S.V, Garratt, R.C, Hasnain, S.S. | | Deposit date: | 2018-02-07 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Molecular recognition and maturation of SOD1 by its evolutionarily destabilised cognate chaperone hCCS.
Plos Biol., 17, 2019
|
|
4RA3
 
 | | Crystal structure of dimeric S33C beta-2 microglobulin mutant in complex with Thioflavin (ThT) at 2.8 Angstrom resolution | | Descriptor: | 2-[4-(dimethylamino)phenyl]-3,6-dimethyl-1,3-benzothiazol-3-ium, Beta-2-microglobulin | | Authors: | Halabelian, L, Bolognesi, M, Ricagno, S. | | Deposit date: | 2014-09-09 | | Release date: | 2015-09-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A covalent homodimer probing early oligomers along amyloid aggregation.
Sci Rep, 5, 2015
|
|
3BIW
 
 | | Crystal structure of the Neuroligin-1/Neurexin-1beta synaptic adhesion complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Arac, D, Boucard, A.A, Ozkan, E, Strop, P, Newell, E, Sudhof, T.C, Brunger, A.T. | | Deposit date: | 2007-12-01 | | Release date: | 2007-12-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structures of Neuroligin-1 and the Neuroligin-1/Neurexin-1beta Complex Reveal Specific Protein-Protein and Protein-Ca(2+) Interactions.
Neuron, 56, 2007
|
|
4GWC
 
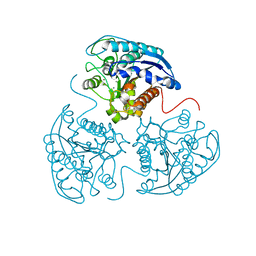 | | Crystal Structure of Mn2+2,Zn2+-Human Arginase I | | Descriptor: | Arginase-1, MANGANESE (II) ION, ZINC ION | | Authors: | D'Antonio, E.L, Hai, Y, Christianson, D.W. | | Deposit date: | 2012-09-01 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and function of non-native metal clusters in human arginase I.
Biochemistry, 51, 2012
|
|
1CWY
 
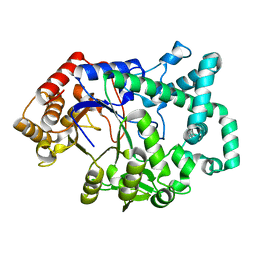 | | CRYSTAL STRUCTURE OF AMYLOMALTASE FROM THERMUS AQUATICUS, A GLYCOSYLTRANSFERASE CATALYSING THE PRODUCTION OF LARGE CYCLIC GLUCANS | | Descriptor: | AMYLOMALTASE | | Authors: | Przylas, I, Tomoo, K, Terada, Y, Takaha, T, Fuji, K, Saenger, W, Straeter, N. | | Deposit date: | 1999-08-27 | | Release date: | 2001-02-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of amylomaltase from thermus aquaticus, a glycosyltransferase catalysing the production of large cyclic glucans.
J.Mol.Biol., 296, 2000
|
|
6FOL
 
 | | Domain II of the human copper chaperone in complex with human Cu,Zn superoxide dismutase | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, Copper chaperone for superoxide dismutase, GLYCEROL, ... | | Authors: | Sala, F.A, Wright, G.S.A, Antonyuk, S.V, Garratt, R.C, Hasnain, S.S. | | Deposit date: | 2018-02-07 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Molecular recognition and maturation of SOD1 by its evolutionarily destabilised cognate chaperone hCCS.
Plos Biol., 17, 2019
|
|
4R9H
 
 | |
6GCT
 
 | | cryo-EM structure of the human neutral amino acid transporter ASCT2 | | Descriptor: | GLUTAMINE, Neutral amino acid transporter B(0) | | Authors: | Garaeva, A.A, Oostergetel, G.T, Gati, C, Guskov, A, Paulino, C, Slotboom, D.J. | | Deposit date: | 2018-04-19 | | Release date: | 2018-06-13 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Cryo-EM structure of the human neutral amino acid transporter ASCT2.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6FP6
 
 | | Complex of human Cu,Zn SOD1 with the human copper chaperone for SOD1 in a compact conformation | | Descriptor: | Copper chaperone for superoxide dismutase, GLYCEROL, PROPANOIC ACID, ... | | Authors: | Sala, F.A, Wright, G.S.A, Antonyuk, S.V, Garratt, R.C, Hasnain, S.S. | | Deposit date: | 2018-02-09 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular recognition and maturation of SOD1 by its evolutionarily destabilised cognate chaperone hCCS.
Plos Biol., 17, 2019
|
|
1XPW
 
 | | Solution NMR Structure of human protein HSPCO34. Northeast Structural Genomics Target HR1958 | | Descriptor: | LOC51668 protein | | Authors: | Ramelot, T.A, Xiao, R, Ma, L.C, Acton, T.B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-10-09 | | Release date: | 2004-11-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Improving NMR protein structure quality by Rosetta refinement: a molecular replacement study.
Proteins, 75, 2009
|
|
3HVX
 
 | |
6FW2
 
 | | Crystal Structure of human mARC1 | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MOLYBDATE ION, Mitochondrial amidoxime-reducing component 1,Endolysin,Mitochondrial amidoxime-reducing component 1, ... | | Authors: | Kubitza, C, Scheidig, A. | | Deposit date: | 2018-03-05 | | Release date: | 2018-10-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of human mARC1 reveals its exceptional position among eukaryotic molybdenum enzymes.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6FWD
 
 | |
4GSM
 
 | |
4GWD
 
 | | Crystal Structure of the Mn2+2,Zn2+-Human Arginase I-ABH Complex | | Descriptor: | 2(S)-AMINO-6-BORONOHEXANOIC ACID, Arginase-1, MANGANESE (II) ION, ... | | Authors: | D'Antonio, E.L, Hai, Y, Christianson, D.W. | | Deposit date: | 2012-09-01 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structure and function of non-native metal clusters in human arginase I.
Biochemistry, 51, 2012
|
|
6DY6
 
 | | Mn(II)-bound structure of the engineered cyt cb562 variant, CH2E | | Descriptor: | HEME C, MANGANESE (II) ION, PENTAETHYLENE GLYCOL, ... | | Authors: | Tezcan, F.A, Rittle, J. | | Deposit date: | 2018-07-01 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An efficient, step-economical strategy for the design of functional metalloproteins.
Nat.Chem., 11, 2019
|
|
6DYE
 
 | | Fe(II)-bound structure of the engineered cyt cb562 variant, CH3 | | Descriptor: | CALCIUM ION, FE (III) ION, HEME C, ... | | Authors: | Tezcan, F.A, Rittle, J. | | Deposit date: | 2018-07-01 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | An efficient, step-economical strategy for the design of functional metalloproteins.
Nat.Chem., 11, 2019
|
|
6DYL
 
 | |
4RAH
 
 | |
3B09
 
 | | Crystal structure of the N-domain of FKBP22 from Shewanella sp. SIB1 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase | | Authors: | Budiman, C, Angkawidjaja, C, Motoike, H, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2011-06-07 | | Release date: | 2012-04-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of N-domain of FKBP22 from Shewanella sp. SIB1: dimer dissociation by disruption of Val-Leu knot
Protein Sci., 20, 2011
|
|
