2QTE
 
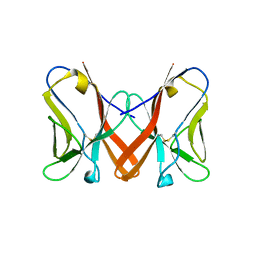 | | Crystal Structure of Novel Immune-Type Receptor 11 Extracellular Fragment Mutant N30D | | Descriptor: | Novel immune-type receptor 11 | | Authors: | Ostrov, D.A, Hernandez Prada, J.A, Haire, R.N, Cannon, J.P, Magis, A.T, Bailey, K.M, Litman, G.W. | | Deposit date: | 2007-08-01 | | Release date: | 2008-09-02 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A bony fish immunological receptor of the NITR multigene family mediates allogeneic recognition.
Immunity, 29, 2008
|
|
2QJD
 
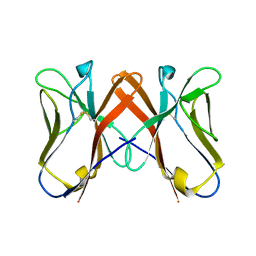 | | Crystal Structure of Novel Immune-Type Receptor 10 Extracellular Fragment Mutant N30D | | Descriptor: | Novel immune-type receptor 10 | | Authors: | Ostrov, D.A, Hernandez Prada, J.A, Haire, R.N, Cannon, J.P, Magis, A.T, Bailey, K.M, Litman, G.W. | | Deposit date: | 2007-07-06 | | Release date: | 2008-06-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | A bony fish immunological receptor of the NITR multigene family mediates allogeneic recognition.
Immunity, 29, 2008
|
|
3HL5
 
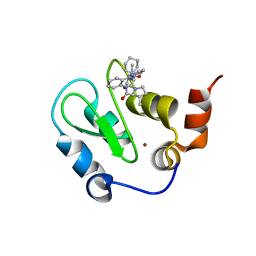 | | Crystal structure of XIAP BIR3 with CS3 | | Descriptor: | (3S)-1-{(2S)-2-cyclohexyl-2-[(N-methyl-L-alanyl)amino]acetyl}-3-methyl-N-(2-pyrimidin-2-ylphenyl)-L-prolinamide, Baculoviral IAP repeat-containing protein 4, ZINC ION | | Authors: | Hymowitz, S.G. | | Deposit date: | 2009-05-26 | | Release date: | 2009-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Antagonism of c-IAP and XIAP proteins is required for efficient induction of cell death by small-molecule IAP antagonists.
Acs Chem.Biol., 4, 2009
|
|
2P3K
 
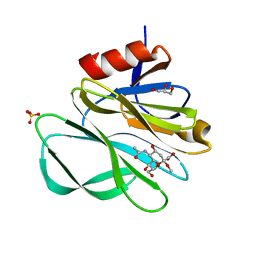 | | Crystal structure of Rhesus rotavirus VP8* at 100K | | Descriptor: | 2-O-methyl-5-N-acetyl-alpha-D-neuraminic acid, GLYCEROL, SULFATE ION, ... | | Authors: | Blanchard, H. | | Deposit date: | 2007-03-09 | | Release date: | 2008-03-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Effects on sialic acid recognition of amino acid mutations in the carbohydrate-binding cleft of the rotavirus spike protein
Glycobiology, 19, 2009
|
|
2RIY
 
 | | B-specific-1,3-galactosyltransferase (GTB)+H-antigen acceptor | | Descriptor: | Glycoprotein-fucosylgalactoside alpha-galactosyltransferase, octyl 2-O-(6-deoxy-alpha-L-galactopyranosyl)-beta-D-galactopyranoside | | Authors: | Evans, S.V, Alfaro, J.A. | | Deposit date: | 2007-10-13 | | Release date: | 2008-02-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | ABO(H) blood group A and B glycosyltransferases recognize substrate via specific conformational changes.
J.Biol.Chem., 283, 2008
|
|
2RJ4
 
 | | B-specific alpha-1,3-galactosyltransferase G176R +UDP+ADA | | Descriptor: | Glycoprotein-fucosylgalactoside alpha-galactosyltransferase, MANGANESE (II) ION, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Evans, S.V, Alfaro, J.A. | | Deposit date: | 2007-10-14 | | Release date: | 2008-02-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | ABO(H) blood group A and B glycosyltransferases recognize substrate via specific conformational changes.
J.Biol.Chem., 283, 2008
|
|
2RIX
 
 | | B-specific-1,3-galactosyltransferase)(GTB) + UDP | | Descriptor: | GLYCEROL, Glycoprotein-fucosylgalactoside alpha-galactosyltransferase, MANGANESE (II) ION, ... | | Authors: | Evans, S.V, Alfaro, J.A. | | Deposit date: | 2007-10-13 | | Release date: | 2008-02-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | ABO(H) blood group A and B glycosyltransferases recognize substrate via specific conformational changes.
J.Biol.Chem., 283, 2008
|
|
2RJ9
 
 | |
1SPU
 
 | | STRUCTURE OF OXIDOREDUCTASE | | Descriptor: | CALCIUM ION, COPPER (II) ION, COPPER AMINE OXIDASE | | Authors: | Wilmot, C.M, Phillips, S.E.V. | | Deposit date: | 1996-11-13 | | Release date: | 1997-03-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Catalytic mechanism of the quinoenzyme amine oxidase from Escherichia coli: exploring the reductive half-reaction.
Biochemistry, 36, 1997
|
|
1TYP
 
 | |
1VH1
 
 | | Crystal structure of CMP-KDO synthetase | | Descriptor: | 3-deoxy-manno-octulosonate cytidylyltransferase | | Authors: | Structural GenomiX | | Deposit date: | 2003-12-01 | | Release date: | 2003-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1VHM
 
 | | Crystal structure of an hypothetical protein | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Protein yebR | | Authors: | Structural GenomiX | | Deposit date: | 2003-12-01 | | Release date: | 2003-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1VHX
 
 | |
1VI6
 
 | | Crystal structure of ribosomal protein S2P | | Descriptor: | 30S ribosomal protein S2P, SODIUM ION | | Authors: | Structural GenomiX | | Deposit date: | 2003-12-01 | | Release date: | 2003-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1VIU
 
 | |
1VHC
 
 | |
1VI8
 
 | |
1VGY
 
 | |
1VHJ
 
 | |
1VGU
 
 | |
1VH0
 
 | | Crystal structure of a hypothetical protein | | Descriptor: | Hypothetical UPF0247 protein SAV0024/SA0023 | | Authors: | Structural GenomiX | | Deposit date: | 2003-11-03 | | Release date: | 2003-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1VHL
 
 | |
1VI1
 
 | |
1VGV
 
 | | Crystal structure of UDP-N-acetylglucosamine_2 epimerase | | Descriptor: | UDP-N-acetylglucosamine 2-epimerase, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | Authors: | Structural GenomiX | | Deposit date: | 2003-11-03 | | Release date: | 2003-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1VIX
 
 | | Crystal structure of a putative peptidase T | | Descriptor: | Peptidase T, SULFATE ION, ZINC ION | | Authors: | Structural GenomiX | | Deposit date: | 2003-12-01 | | Release date: | 2003-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
