3K5R
 
 | |
3K6D
 
 | |
5XWJ
 
 | | Crystal Structure of Porcine pancreatic trypsin with tripeptide inhibitor, TRE, at pH 7 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Acetylated-THR-ARG-GLU Inhibitor, CALCIUM ION, ... | | Authors: | Saikhedkar, N.S, Bhoite, A.S, Giri, A.P, Kulkarni, K.A. | | Deposit date: | 2017-06-29 | | Release date: | 2018-03-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Tripeptides derived from reactive centre loop of potato type II protease inhibitors preferentially inhibit midgut proteases of Helicoverpa armigera.
Insect Biochem. Mol. Biol., 95, 2018
|
|
3JUS
 
 | | Crystal structure of human lanosterol 14alpha-demethylase (CYP51) in complex with econazole | | Descriptor: | 1-[(2R)-2-[(4-chlorobenzyl)oxy]-2-(2,4-dichlorophenyl)ethyl]-1H-imidazole, 1-[(2S)-2-[(4-CHLOROBENZYL)OXY]-2-(2,4-DICHLOROPHENYL)ETHYL]-1H-IMIDAZOLE, Cycloheptakis-(1-4)-(alpha-D-glucopyranose), ... | | Authors: | Strushkevich, N, MacKenzie, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Usanov, S.A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-15 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of human CYP51 inhibition by antifungal azoles.
J. Mol. Biol., 397, 2010
|
|
5XC3
 
 | |
3JVR
 
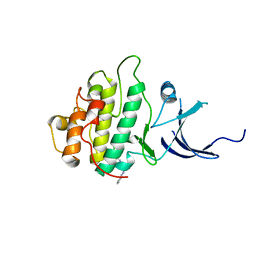 | | Characterization of the Chk1 allosteric inhibitor binding site | | Descriptor: | (1S)-1-(1H-benzimidazol-2-yl)ethyl (3,4-dichlorophenyl)carbamate, Serine/threonine-protein kinase Chk1 | | Authors: | Chen, P. | | Deposit date: | 2009-09-17 | | Release date: | 2009-10-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Characterization of the CHK1 allosteric inhibitor binding site.
Biochemistry, 48, 2009
|
|
5XEC
 
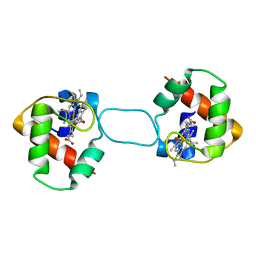 | | Heterodimer constructed from PA cyt c551-HT cyt c552 and HT cyt c552-PA cyt c551 chimeric proteins | | Descriptor: | Cytochrome c-551,Cytochrome c-552, Cytochrome c-552,Cytochrome c-551, HEME C | | Authors: | Zhang, M, Nakanishi, T, Yamanaka, M, Nagao, S, Yanagisawa, S, Shomura, Y, Shibata, N, Ogura, T, Higuchi, Y, Hirota, S. | | Deposit date: | 2017-04-04 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Rational Design of Domain-Swapping-Based c-Type Cytochrome Heterodimers by Using Chimeric Proteins.
Chembiochem, 18, 2017
|
|
3JYA
 
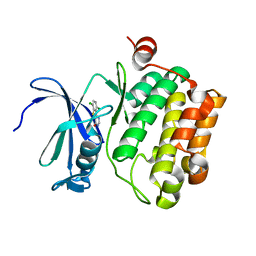 | | Discovery of 3H-benzo[4,5]thieno[3,2-d]pyrimidin-4-ones as Potent, Highly Selective and Orally Bioavailable Pim Kinases Inhibitors | | Descriptor: | 6,9-dichloro[1]benzothieno[3,2-d]pyrimidin-4(3H)-one, Proto-oncogene serine/threonine-protein kinase Pim-1 | | Authors: | Stoll, V.S. | | Deposit date: | 2009-09-21 | | Release date: | 2009-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of 3H-benzo[4,5]thieno[3,2-d]pyrimidin-4-ones as Potent, Highly Selective and Orally Bioavailable Pim Kinases Inhibitors
TO BE PUBLISHED
|
|
5XG9
 
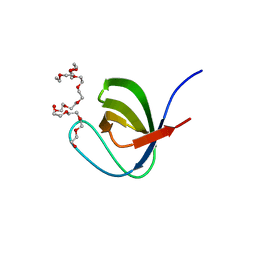 | | Crystal Structure of PEG-bound SH3 domain of Myosin IB from Entamoeba histolytica | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2,5,8,11,14,17,20,23,26,29,32,35,38,41,44,47,50,53,56,59,62,65,68,71,74,77,80-HEPTACOSAOXADOOCTACONTAN-82-OL, PENTAETHYLENE GLYCOL, ... | | Authors: | Gautam, G, Gourinath, S. | | Deposit date: | 2017-04-13 | | Release date: | 2017-08-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of the PEG-bound SH3 domain of myosin IB from Entamoeba histolytica reveals its mode of ligand recognition
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5XJP
 
 | | Crystal structure of response regulator AdeR receiver domain with Mg | | Descriptor: | AdeR, MAGNESIUM ION | | Authors: | Wen, Y. | | Deposit date: | 2017-05-03 | | Release date: | 2017-08-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.597 Å) | | Cite: | Mechanistic insight into how multidrug resistant Acinetobacter baumannii response regulator AdeR recognizes an intercistronic region.
Nucleic Acids Res., 45, 2017
|
|
3K2M
 
 | |
3K5D
 
 | | Crystal Structure of BACE-1 in complex with AHM178 | | Descriptor: | Beta-secretase 1, N-acetyl-L-leucyl-N-[(4S,5S,7R)-8-(butylamino)-5-hydroxy-2,7-dimethyl-8-oxooctan-4-yl]-L-methioninamide | | Authors: | Rondeau, J.-M. | | Deposit date: | 2009-10-07 | | Release date: | 2010-05-05 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-based design and synthesis of novel P2/P3 modified, non-peptidic beta-secretase (BACE-1) inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3K3U
 
 | |
3K6F
 
 | |
3K5G
 
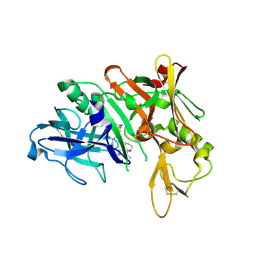 | | Human bace-1 complex with bjc060 | | Descriptor: | (1R,3S)-N-[(1S,2R)-1-benzyl-2-hydroxy-3-{[3-(1-methylethyl)benzyl]amino}propyl]-3-[1-methyl-1-(2-oxopiperidin-1-yl)ethyl]cyclohexanecarboxamide, Beta-secretase 1 | | Authors: | Rondeau, J.-M. | | Deposit date: | 2009-10-07 | | Release date: | 2010-05-05 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design and synthesis of novel P2/P3 modified, non-peptidic beta-secretase (BACE-1) inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3K9V
 
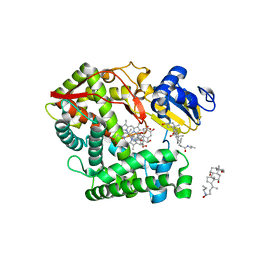 | | Crystal structure of rat mitochondrial P450 24A1 S57D in complex with CHAPS | | Descriptor: | 1,25-dihydroxyvitamin D(3) 24-hydroxylase, mitochondrial, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, ... | | Authors: | Annalora, A.J, Goodin, D.B, Hong, W, Zhang, Q, Johnson, E.F, Stout, C.D. | | Deposit date: | 2009-10-16 | | Release date: | 2009-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of CYP24A1, a mitochondrial cytochrome P450 involved in vitamin D metabolism.
J.Mol.Biol., 396, 2010
|
|
3J8J
 
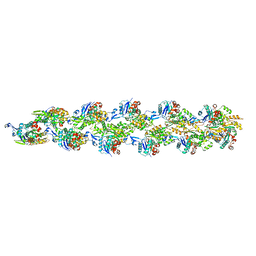 | | Tilted state of actin, T1 | | Descriptor: | Actin, alpha skeletal muscle | | Authors: | Galkin, V.E, Orlova, A, Vos, M.R, Schroder, G.F, Egelman, E.H. | | Deposit date: | 2014-11-07 | | Release date: | 2015-01-14 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Near-atomic resolution for one state of f-actin.
Structure, 23, 2015
|
|
5XVF
 
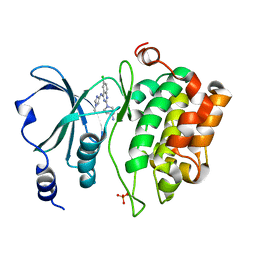 | | Crystal Structure of PAK4 in complex with inhibitor CZH062 | | Descriptor: | 2-(4-azanylpiperidin-1-yl)-6-chloranyl-N-(1-methylimidazol-4-yl)quinazolin-4-amine, Serine/threonine-protein kinase PAK 4 | | Authors: | Zhao, F, Li, H. | | Deposit date: | 2017-06-27 | | Release date: | 2018-02-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.655 Å) | | Cite: | Structure-Based Design of 6-Chloro-4-aminoquinazoline-2-carboxamide Derivatives as Potent and Selective p21-Activated Kinase 4 (PAK4) Inhibitors.
J. Med. Chem., 61, 2018
|
|
5XWL
 
 | | Crystal Structure of Porcine pancreatic trypsin with tripeptide inhibitor, TRE, at pH 10 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Acetylated-THR-ARG-GLU Inhibitor, CALCIUM ION, ... | | Authors: | Saikhedkar, N.S, Bhoite, A.S, Giri, A.P, Kulkarni, K.A. | | Deposit date: | 2017-06-29 | | Release date: | 2018-03-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Tripeptides derived from reactive centre loop of potato type II protease inhibitors preferentially inhibit midgut proteases of Helicoverpa armigera.
Insect Biochem. Mol. Biol., 95, 2018
|
|
3KA2
 
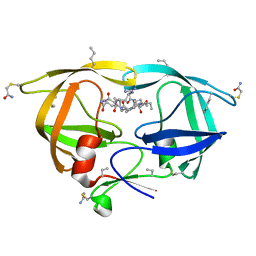 | |
5X83
 
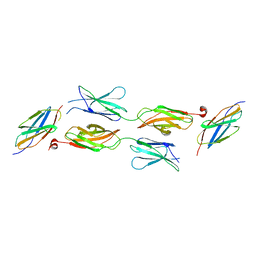 | | Structure of DCC FN456 domains | | Descriptor: | Netrin receptor DCC | | Authors: | Finci, F.I, Xiao, J, Wang, J. | | Deposit date: | 2017-02-28 | | Release date: | 2017-09-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.997 Å) | | Cite: | Structure of unliganded membrane-proximal domains FN4-FN5-FN6 of DCC
Protein Cell, 8, 2017
|
|
5XG4
 
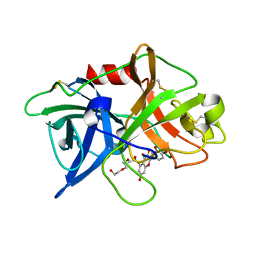 | | Crystal structure of uPA in complex with quercetin | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 3,5,7,3',4'-PENTAHYDROXYFLAVONE, Urokinase-type plasminogen activator | | Authors: | Jiang, L, Huang, M. | | Deposit date: | 2017-04-11 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A structural mechanism of flavonoids in inhibiting serine proteases
Food Funct, 8, 2017
|
|
3JVY
 
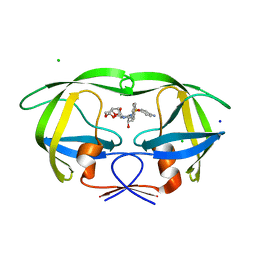 | | HIV-1 Protease Mutant G86A with DARUNAVIR | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, CHLORIDE ION, Gag-Pol polyprotein, ... | | Authors: | Tie, Y, Weber, I.T. | | Deposit date: | 2009-09-17 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Highly conserved glycine 86 and arginine 87 residues contribute differently to the structure and activity of the mature HIV-1 protease
Proteins, 78, 2009
|
|
3JY0
 
 | |
5XHJ
 
 | | Crystal Structure of P450BM3 with 5-Cyclohexylvaleroyl-L-Tryptophan | | Descriptor: | 5-cyclohexylpentanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Suzuki, K, Shoji, O, Stanfield, J.K, Kasai, C, Sugimoto, H, Shiro, Y, Watanabe, Y. | | Deposit date: | 2017-04-21 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Control of stereoselectivity of benzylic hydroxylation catalysed by wild-type cytochrome P450BM3 using decoy molecules
CATALYSIS SCIENCE AND TECHNOLOGY, 7, 2017
|
|
