7XMF
 
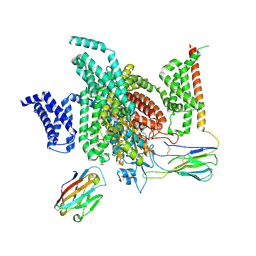 | | Cryo-EM structure of human NaV1.7/beta1/beta2-Nav1.7-IN2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[[4-[3-(4-fluoranyl-2-methyl-phenoxy)azetidin-1-yl]pyrimidin-2-yl]amino]-~{N}-methyl-benzamide, ... | | 著者 | Zhang, J.T, Jiang, D.H. | | 登録日 | 2022-04-25 | | 公開日 | 2022-11-30 | | 最終更新日 | 2022-12-28 | | 実験手法 | ELECTRON MICROSCOPY (3.07 Å) | | 主引用文献 | Structural basis for Na V 1.7 inhibition by pore blockers.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7XM9
 
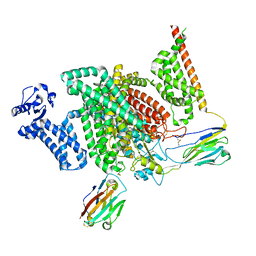 | | Cryo-EM structure of human NaV1.7/beta1/beta2-XEN907 | | 分子名称: | (7~{R})-1'-pentylspiro[6~{H}-furo[3,2-f][1,3]benzodioxole-7,3'-indole]-2'-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | zhang, J.T, Jiang, D.H. | | 登録日 | 2022-04-25 | | 公開日 | 2022-11-30 | | 最終更新日 | 2022-12-28 | | 実験手法 | ELECTRON MICROSCOPY (3.22 Å) | | 主引用文献 | Structural basis for Na V 1.7 inhibition by pore blockers.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7XJ3
 
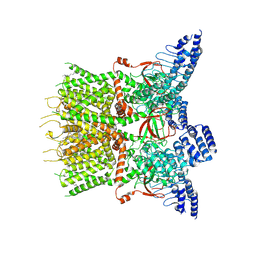 | | Structure of human TRPV3 | | 分子名称: | [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate, fusion of transient receptor potential cation channel subfamily V member 3 and 3C-GFP | | 著者 | Fan, J, Yue, Z, Jiang, D, Lei, X. | | 登録日 | 2022-04-14 | | 公開日 | 2022-11-09 | | 最終更新日 | 2023-01-11 | | 実験手法 | ELECTRON MICROSCOPY (3.54 Å) | | 主引用文献 | Structural basis of TRPV3 inhibition by an antagonist.
Nat.Chem.Biol., 19, 2023
|
|
7XJ2
 
 | | Structure of human TRPV3_G573S in complex with Trpvicin in C4 symmetry | | 分子名称: | Fusion protein of Transient receptor potential cation channel subfamily V member 3 and 3C-GFP, N-[5-[2-(2-cyanopropan-2-yl)pyridin-4-yl]-4-(trifluoromethyl)-1,3-thiazol-2-yl]-4,6-dimethoxy-pyrimidine-5-carboxamide | | 著者 | Fan, J, Yue, Z, Jiang, D, Lei, X. | | 登録日 | 2022-04-14 | | 公開日 | 2022-11-09 | | 最終更新日 | 2023-01-11 | | 実験手法 | ELECTRON MICROSCOPY (3.64 Å) | | 主引用文献 | Structural basis of TRPV3 inhibition by an antagonist.
Nat.Chem.Biol., 19, 2023
|
|
7XJ1
 
 | | Structure of human TRPV3_G573S in complex with Trpvicin in C2 symmetry | | 分子名称: | Fusion protein of Transient receptor potential cation channel subfamily V member 3 and 3C-GFP, N-[5-[2-(2-cyanopropan-2-yl)pyridin-4-yl]-4-(trifluoromethyl)-1,3-thiazol-2-yl]-4,6-dimethoxy-pyrimidine-5-carboxamide, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate | | 著者 | Fan, J, Yue, Z, Jiang, D, Lei, X. | | 登録日 | 2022-04-14 | | 公開日 | 2022-11-09 | | 最終更新日 | 2023-01-11 | | 実験手法 | ELECTRON MICROSCOPY (2.93 Å) | | 主引用文献 | Structural basis of TRPV3 inhibition by an antagonist.
Nat.Chem.Biol., 19, 2023
|
|
7XJ0
 
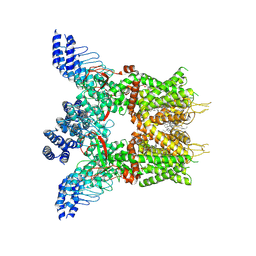 | | Structure of human TRPV3 in complex with Trpvicin | | 分子名称: | Fusion protein of Transient receptor potential cation channel subfamily V member 3 and 3C-GFP, N-[5-[2-(2-cyanopropan-2-yl)pyridin-4-yl]-4-(trifluoromethyl)-1,3-thiazol-2-yl]-4,6-dimethoxy-pyrimidine-5-carboxamide, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate | | 著者 | Fan, J, Yue, Z, Jiang, D, Lei, X. | | 登録日 | 2022-04-14 | | 公開日 | 2022-11-09 | | 最終更新日 | 2023-01-11 | | 実験手法 | ELECTRON MICROSCOPY (2.53 Å) | | 主引用文献 | Structural basis of TRPV3 inhibition by an antagonist.
Nat.Chem.Biol., 19, 2023
|
|
7X5V
 
 | |
7VCM
 
 | | crystal structure of GINKO1 | | 分子名称: | Green fluorescent protein,Potassium binding protein Kbp,Green fluorescent protein, POTASSIUM ION | | 著者 | Wen, Y, Campbell, R.E, Lemieux, M.J. | | 登録日 | 2021-09-03 | | 公開日 | 2022-07-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | A sensitive and specific genetically-encoded potassium ion biosensor for in vivo applications across the tree of life.
Plos Biol., 20, 2022
|
|
7V80
 
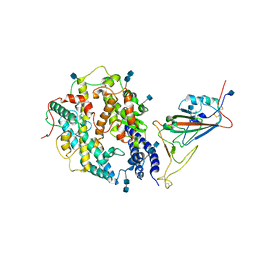 | | Local refinement of SARS-CoV-2 S-Beta variant (B.1.351) RBD and Angiotensin-converting enzyme 2 (ACE2) ectodomain | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2,Green fluorescent protein, ... | | 著者 | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | 登録日 | 2021-08-22 | | 公開日 | 2021-10-06 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Local refinement of SARS-CoV-2 S-Beta variant (B.1.351) RBD and Angiotensin-converting enzyme 2 (ACE2) ectodomain
To Be Published
|
|
7V0V
 
 | | GFP Nanobody NMR Structure | | 分子名称: | Anti-GFP Nanobody | | 著者 | Mueller, G.A. | | 登録日 | 2022-05-11 | | 公開日 | 2022-06-08 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Nanobody Paratope Ensembles in Solution Characterized by MD Simulations and NMR.
Int J Mol Sci, 23, 2022
|
|
7UK4
 
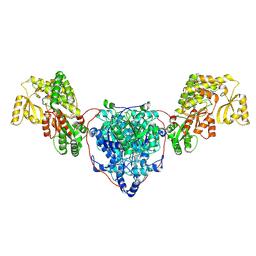 | | KS-AT di-domain of mycobacterial Pks13 with endogenous KS ligand bound | | 分子名称: | Polyketide synthase PKS13, UNKNOWN LIGAND | | 著者 | Kim, S.K, Dickinson, M.S, Finer-Moore, J.S, Rosenberg, O.S, Stroud, R.M. | | 登録日 | 2022-03-31 | | 公開日 | 2023-02-15 | | 最終更新日 | 2023-03-29 | | 実験手法 | ELECTRON MICROSCOPY (1.94 Å) | | 主引用文献 | Structure and dynamics of the essential endogenous mycobacterial polyketide synthase Pks13.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7UJ0
 
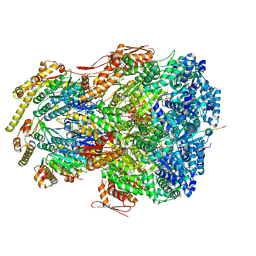 | | ClpAP complex bound to ClpS N-terminal extension, class IIIb | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ATP-dependent Clp protease adapter protein ClpS, ... | | 著者 | Kim, S, Fei, X, Sauer, R.T, Baker, T.A. | | 登録日 | 2022-03-29 | | 公開日 | 2022-11-09 | | 最終更新日 | 2022-11-30 | | 実験手法 | ELECTRON MICROSCOPY (3.26 Å) | | 主引用文献 | AAA+ protease-adaptor structures reveal altered conformations and ring specialization.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7UIZ
 
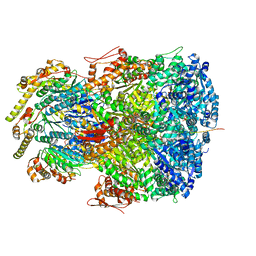 | | ClpAP complex bound to ClpS N-terminal extension, class IIc | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ATP-dependent Clp protease adapter protein ClpS, ... | | 著者 | Kim, S, Fei, X, Sauer, R.T, Baker, T.A. | | 登録日 | 2022-03-29 | | 公開日 | 2022-11-09 | | 最終更新日 | 2022-11-30 | | 実験手法 | ELECTRON MICROSCOPY (3.24 Å) | | 主引用文献 | AAA+ protease-adaptor structures reveal altered conformations and ring specialization.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7UIY
 
 | | ClpAP complex bound to ClpS N-terminal extension, class IIIa | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ATP-dependent Clp protease adapter protein ClpS, ... | | 著者 | Kim, S, Fei, X, Sauer, R.T, Baker, T.A. | | 登録日 | 2022-03-29 | | 公開日 | 2022-10-26 | | 最終更新日 | 2022-11-30 | | 実験手法 | ELECTRON MICROSCOPY (3.22 Å) | | 主引用文献 | AAA+ protease-adaptor structures reveal altered conformations and ring specialization.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7UIX
 
 | | ClpAP complex bound to ClpS N-terminal extension, class I | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ATP-dependent Clp protease adapter protein ClpS, ... | | 著者 | Kim, S, Fei, X, Sauer, R.T, Baker, T.A. | | 登録日 | 2022-03-29 | | 公開日 | 2022-11-09 | | 最終更新日 | 2022-11-30 | | 実験手法 | ELECTRON MICROSCOPY (3.24 Å) | | 主引用文献 | AAA+ protease-adaptor structures reveal altered conformations and ring specialization.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7UIW
 
 | | ClpAP complex bound to ClpS N-terminal extension, class IIb | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ATP-dependent Clp protease adapter protein ClpS, ... | | 著者 | Kim, S, Fei, X, Sauer, R.T, Baker, T.A. | | 登録日 | 2022-03-29 | | 公開日 | 2022-11-09 | | 最終更新日 | 2022-11-30 | | 実験手法 | ELECTRON MICROSCOPY (3.33 Å) | | 主引用文献 | AAA+ protease-adaptor structures reveal altered conformations and ring specialization.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7UIV
 
 | | ClpAP complex bound to ClpS N-terminal extension, class IIa | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ATP-dependent Clp protease adapter protein ClpS, ... | | 著者 | Kim, S, Fei, X, Sauer, R.T, Baker, T.A. | | 登録日 | 2022-03-29 | | 公開日 | 2022-11-09 | | 最終更新日 | 2022-11-30 | | 実験手法 | ELECTRON MICROSCOPY (3.38 Å) | | 主引用文献 | AAA+ protease-adaptor structures reveal altered conformations and ring specialization.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7UGT
 
 | |
7UGS
 
 | |
7UGR
 
 | | Crystal structure of hyperfolder YFP | | 分子名称: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Hyperfolder yellow fluorescent protein, ... | | 著者 | Campbell, B.C, Liu, C.F, Petsko, G.A. | | 登録日 | 2022-03-25 | | 公開日 | 2022-10-26 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.74 Å) | | 主引用文献 | Chemically stable fluorescent proteins for advanced microscopy.
Nat.Methods, 19, 2022
|
|
7TSV
 
 | |
7TSU
 
 | |
7TSS
 
 | |
7TSR
 
 | |
7SSZ
 
 | |
