6BEU
 
 | | Solution structure of de novo macrocycle design14_ss | | Descriptor: | (DCY)N(DVA)(DPR)DVYC(DPR)(DSG)KY(DVA)(DPR) | | Authors: | Shortridge, M.D, Hosseinzadeh, P, Pardo-Avila, F, Varani, G, Baker, D. | | Deposit date: | 2017-10-25 | | Release date: | 2017-12-27 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Comprehensive computational design of ordered peptide macrocycles.
Science, 358, 2017
|
|
6BEO
 
 | | Solution structure of de novo macrocycle design9.1 | | Descriptor: | (DPR)PY(DHI)PKDL(DGN) | | Authors: | Shortridge, M.D, Hosseinzadeh, P, Pardo-Avila, F, Varani, G, Baker, D. | | Deposit date: | 2017-10-25 | | Release date: | 2017-12-27 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Comprehensive computational design of ordered peptide macrocycles.
Science, 358, 2017
|
|
6BE9
 
 | | Solution structure of de novo macrocycle design7.1 | | Descriptor: | T(DLY)NDT(DSG)(DPR) | | Authors: | Shortridge, M.D, Hosseinzadeh, P, Pardo-Avila, F, Varani, G, Baker, B. | | Deposit date: | 2017-10-24 | | Release date: | 2017-12-27 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Comprehensive computational design of ordered peptide macrocycles.
Science, 358, 2017
|
|
8UI2
 
 | | T33-ml28 - Designed Tetrahedral Protein Cage Using Machine Learning Algorithms | | Descriptor: | T33-ml28-redesigned-CutA-fold, T33-ml28-redesigned-tandem-BMC-T-fold | | Authors: | Castells-Graells, R, Meador, K, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2023-10-09 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | A suite of designed protein cages using machine learning and protein fragment-based protocols.
Structure, 32, 2024
|
|
8UKM
 
 | | T33-ml30 - Designed Tetrahedral Protein Cage Using Machine Learning Algorithms | | Descriptor: | T33-ml30-redesigned-4-OT-fold, T33-ml30-redesigned-tandem-BMC-T-fold | | Authors: | Castells-Graells, R, Meador, K, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2023-10-14 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | A suite of designed protein cages using machine learning and protein fragment-based protocols.
Structure, 32, 2024
|
|
8UN1
 
 | | T33-ml23 Assembly Intermediate - Designed Tetrahedral Protein Cage Using Machine Learning Algorithms | | Descriptor: | T33-ml23-redesigned-CutA-fold, T33-ml23-redesigned-tandem-BMC-T-fold | | Authors: | Castells-Graells, R, Meador, K, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2023-10-18 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | A suite of designed protein cages using machine learning and protein fragment-based protocols.
Structure, 32, 2024
|
|
8UMR
 
 | | T33-ml35 Assembly Intermediate - Designed Tetrahedral Protein Cage Using Machine Learning Algorithms | | Descriptor: | T33-ml35-redesigned-CutA-fold, T33-ml35-redesigned-TPR-domain-fold | | Authors: | Castells-Graells, R, Meador, K, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2023-10-18 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.42 Å) | | Cite: | A suite of designed protein cages using machine learning and protein fragment-based protocols.
Structure, 32, 2024
|
|
4U3H
 
 | | Crystal structure of FN3con | | Descriptor: | FN3con | | Authors: | Porebski, B.T, McGowan, S, Buckle, A.M. | | Deposit date: | 2014-07-21 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural and dynamic properties that govern the stability of an engineered fibronectin type III domain.
Protein Eng.Des.Sel., 28, 2015
|
|
1CIB
 
 | | STRUCTURE OF ADENYLOSUCCINATE SYNTHETASE FROM E. COLI COMPLEXED WITH GDP, IMP, HADACIDIN, AND NO3 | | Descriptor: | ADENYLOSUCCINATE SYNTHETASE, GUANOSINE-5'-DIPHOSPHATE, HADACIDIN, ... | | Authors: | Hou, Z, Cashel, M, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 1999-03-31 | | Release date: | 2000-04-05 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Effectors of the stringent response target the active site of Escherichia coli adenylosuccinate synthetase.
J.Biol.Chem., 274, 1999
|
|
6REG
 
 | | Crystal structure of Pizza6-S with Zn2+ | | Descriptor: | GLYCEROL, Pizza6-S, ZINC ION | | Authors: | Noguchi, H, Clarke, D.E, Gryspeerdt, J.L, Feyter, S.D, Voet, A.R.D. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Artificial beta-propeller protein-based hydrolases.
Chem.Commun.(Camb.), 55, 2019
|
|
6REK
 
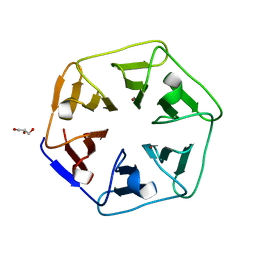 | | Crystal structure of Pizza6-SH with Cu2+ | | Descriptor: | COPPER (II) ION, GLYCEROL, Pizza6-SH | | Authors: | Noguchi, H, Clarke, D.E, Gryspeerdt, J.L, Feyter, S.D, Voet, A.R.D. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Artificial beta-propeller protein-based hydrolases.
Chem.Commun.(Camb.), 55, 2019
|
|
1DJ2
 
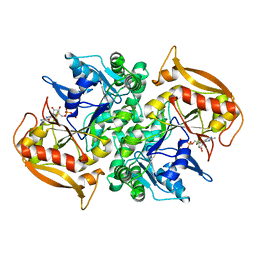 | | STRUCTURES OF ADENYLOSUCCINATE SYNTHETASE FROM TRITICUM AESTIVUM AND ARABIDOPSIS THALIANA | | Descriptor: | ADENYLOSUCCINATE SYNTHETASE, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Prade, L, Cowan-Jacob, S.W, Chemla, P, Potter, S, Ward, E, Fonne-Pfister, R. | | Deposit date: | 1999-12-01 | | Release date: | 2000-03-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of adenylosuccinate synthetase from Triticum aestivum and Arabidopsis thaliana.
J.Mol.Biol., 296, 2000
|
|
1DY3
 
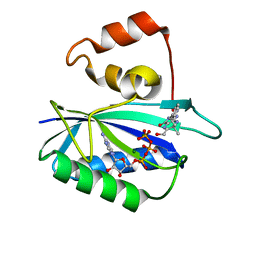 | | Ternary complex of 7,8-dihydro-6-hydroxymethylpterinpyrophosphokinase from Escherichia coli with ATP and a substrate analogue. | | Descriptor: | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, 7,8-DIHYDRO-6-HYDROXYMETHYL-7-METHYL-7-[2-PHENYLETHYL]-PTERIN, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Stammers, D.K, Achari, A, Somers, D.O, Bryant, P.K, Rosemond, J, Scott, D.L, Champness, J.N. | | Deposit date: | 2000-01-21 | | Release date: | 2000-08-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2.0A X-Ray Structure of the Ternary Complex of 7,8-Dihydro-6-Hydroxymethylpterinpyrophosphokinase from Escherichia Coli with ATP and a Substrate Analogue
FEBS Lett., 456, 1999
|
|
1EIB
 
 | | CRYSTAL STRUCTURE OF CHITINASE A MUTANT D313A COMPLEXED WITH OCTA-N-ACETYLCHITOOCTAOSE (NAG)8. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHITINASE A | | Authors: | Papanikolau, Y, Prag, G, Tavlas, G, Vorgias, C.E, Oppenheim, A.B, Petratos, K. | | Deposit date: | 2000-02-25 | | Release date: | 2001-02-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High resolution structural analyses of mutant chitinase A complexes with substrates provide new insight into the mechanism of catalysis.
Biochemistry, 40, 2001
|
|
8FG6
 
 | |
1EZ1
 
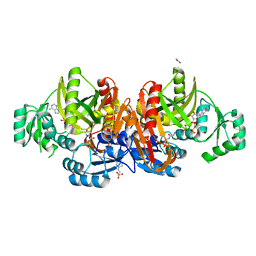 | | STRUCTURE OF ESCHERICHIA COLI PURT-ENCODED GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE COMPLEXED WITH MG, AMPPNP, AND GAR | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, ACETATE ION, GLYCINAMIDE RIBONUCLEOTIDE, ... | | Authors: | Thoden, J.B, Firestine, S, Nixon, A, Benkovic, S.J, Holden, H.M. | | Deposit date: | 2000-05-09 | | Release date: | 2000-08-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Molecular structure of Escherichia coli PurT-encoded glycinamide ribonucleotide transformylase.
Biochemistry, 39, 2000
|
|
1EHN
 
 | | CRYSTAL STRUCTURE OF CHITINASE A MUTANT E315Q COMPLEXED WITH OCTA-N-ACETYLCHITOOCTAOSE (NAG)8. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHITINASE A | | Authors: | Papanikolau, Y, Prag, G, Tavlas, G, Vorgias, C.E, Oppenheim, A.B, Petratos, K. | | Deposit date: | 2000-02-22 | | Release date: | 2001-02-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High resolution structural analyses of mutant chitinase A complexes with substrates provide new insight into the mechanism of catalysis.
Biochemistry, 40, 2001
|
|
6REH
 
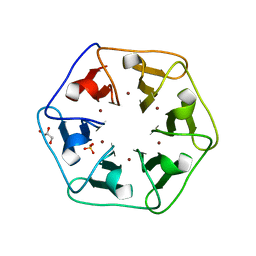 | | Crystal structure of Pizza6-S with Cu2+ | | Descriptor: | COPPER (II) ION, GLYCEROL, Pizza6-S, ... | | Authors: | Noguchi, H, Clarke, D.E, Gryspeerdt, J.L, Feyter, S.D, Voet, A.R.D. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Artificial beta-propeller protein-based hydrolases.
Chem.Commun.(Camb.), 55, 2019
|
|
6REJ
 
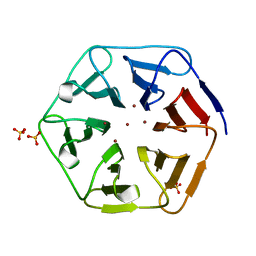 | | Crystal structure of Pizza6-SH with Zn2+ | | Descriptor: | Pizza6-SH, SULFATE ION, ZINC ION | | Authors: | Noguchi, H, Clarke, D.E, Gryspeerdt, J.L, Feyter, S.D, Voet, A.R.D. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Artificial beta-propeller protein-based hydrolases.
Chem.Commun.(Camb.), 55, 2019
|
|
6REN
 
 | | Crystal structure of 3fPizza6-SH with Zn2+ | | Descriptor: | 3fPizza6-SH, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Noguchi, H, Clarke, D.E, Gryspeerdt, J.L, Feyter, S.D, Voet, A.R.D. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Artificial beta-propeller protein-based hydrolases.
Chem.Commun.(Camb.), 55, 2019
|
|
1EYZ
 
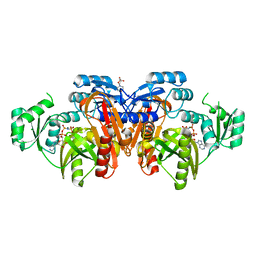 | | STRUCTURE OF ESCHERICHIA COLI PURT-ENCODED GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE COMPLEXED WITH MG AND AMPPNP | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Thoden, J.B, Firestine, S, Nixon, A, Benkovic, S.J, Holden, H.M. | | Deposit date: | 2000-05-09 | | Release date: | 2000-08-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Molecular structure of Escherichia coli PurT-encoded glycinamide ribonucleotide transformylase.
Biochemistry, 39, 2000
|
|
6B6H
 
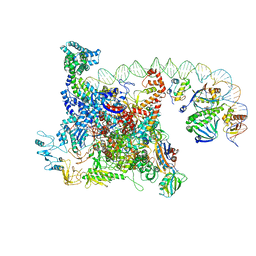 | | The cryo-EM structure of a bacterial class I transcription activation complex | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Liu, B, Hong, C, Huang, R, Yu, Z, Steitz, T.A. | | Deposit date: | 2017-10-02 | | Release date: | 2017-11-15 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis of bacterial transcription activation.
Science, 358, 2017
|
|
7BO9
 
 | |
7BO8
 
 | | A hexameric de novo coiled-coil assembly: CC-Type2-(VaYd)4-Y3F-W19(BrPhe)-Y24F. | | Descriptor: | 1,2-ETHANEDIOL, CC-Type2-(VaYd)4-Y3F-W19(BrPhe)-Y24F, OXAMIC ACID | | Authors: | Rhys, G.G, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2021-01-24 | | Release date: | 2021-05-19 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | How Coiled-Coil Assemblies Accommodate Multiple Aromatic Residues.
Biomacromolecules, 22, 2021
|
|
7BOA
 
 | |
