7OL2
 
 | | Crystal structure of mouse contactin 1 immunoglobulin domains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Contactin-1, ... | | Authors: | Chataigner, L.M.P, Janssen, B.J.C. | | Deposit date: | 2021-05-19 | | Release date: | 2022-12-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.89 Å) | | Cite: | Structural insights into the contactin 1 - neurofascin 155 adhesion complex.
Nat Commun, 13, 2022
|
|
8KBH
 
 | |
6MVF
 
 | |
5CIS
 
 | | The CUB1-EGF-CUB2 domains of rat MBL-associated serine protease-2 (MASP-2) bound to Ca2+ | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Mannan-binding lectin serine peptidase 2 | | Authors: | Nan, R, Furze, C.M, Wright, D.W, Gor, J, Wallis, R, Perkins, S.J. | | Deposit date: | 2015-07-13 | | Release date: | 2017-01-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Flexibility in Mannan-Binding Lectin-Associated Serine Proteases-1 and -2 Provides Insight on Lectin Pathway Activation.
Structure, 25, 2017
|
|
7ON6
 
 | | Crystal structure of the computationally designed SAKe6AE protein | | Descriptor: | SAKe6AE, SULFATE ION | | Authors: | Wouters, S.M.L, Noguchi, H, Clarke, D.E, Voet, A.R.D, De Feyter, S. | | Deposit date: | 2021-05-25 | | Release date: | 2022-12-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | SAKe: Computationally Designed Modular Protein Building Blocks for Macromolecular Assemblies
To be published
|
|
8RZE
 
 | | SARS-CoV-2 nsp16-nsp10 in complex with SAM derivative inhibitor 10 | | Descriptor: | 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-[[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanylmethyl]-5-pyridin-3-yl-benzoic acid, ... | | Authors: | Kalnins, G. | | Deposit date: | 2024-02-12 | | Release date: | 2024-02-21 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for inhibition of the SARS-CoV-2 nsp16 by substrate-based dual site inhibitors.
Chemmedchem, 2024
|
|
6N3G
 
 | | Crystal structure of histone lysine methyltransferase SmyD2 in complex with polyethylene glycol | | Descriptor: | DODECAETHYLENE GLYCOL, ETHANOL, N-lysine methyltransferase SMYD2, ... | | Authors: | Perry, E, Spellmon, N, Brunzelle, J, Yang, Z. | | Deposit date: | 2018-11-15 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Crystal structure of histone lysine methyltransferase SmyD2 in complex with polyethylene glycol
To be Published
|
|
7ON8
 
 | |
6N3S
 
 | | Crystal structure of apo-cruzain | | Descriptor: | 1,2-ETHANEDIOL, Cruzipain, PHOSPHATE ION | | Authors: | Silva, E.B, Dall, E, Rodrigues, F.T.G, Ferreira, R.S, Brandstetter, H. | | Deposit date: | 2018-11-16 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.193 Å) | | Cite: | Cruzain structures: apocruzain and cruzain bound to S-methyl thiomethanesulfonate and implications for drug design.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
8RQU
 
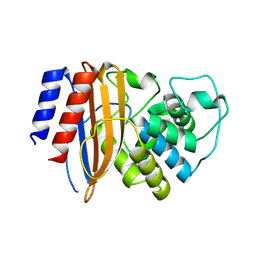 | | Structure of TEM1 beta-lactamase variant 70.a | | Descriptor: | Beta-lactamase TEM-1, MAGNESIUM ION | | Authors: | Napier, E, Fram, B.F, Gauthier, N.P, Sander, C, Khan, A.R. | | Deposit date: | 2024-01-19 | | Release date: | 2024-02-14 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Simultaneous enhancement of multiple functional properties using evolution-informed protein design.
Nat Commun, 15, 2024
|
|
7ONA
 
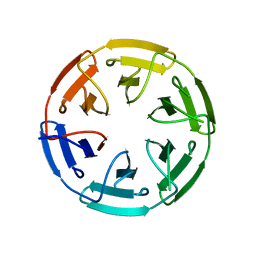 | | Crystal structure of the computationally designed SAKe6AC protein | | Descriptor: | CALCIUM ION, SAKe6AC | | Authors: | Wouters, S.M.L, Noguchi, H, Velpula, G, Clarke, D.E, Voet, A.R.D, De Feyter, S. | | Deposit date: | 2021-05-25 | | Release date: | 2022-12-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | SAKe: Computationally Designed Modular Protein Building Blocks for Macromolecular Assemblies
To be published
|
|
7ONE
 
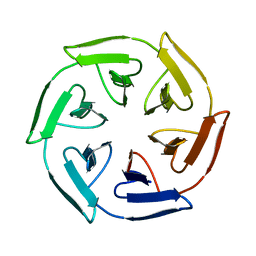 | | Crystal structure of the self-assembled SAKe6BE designer protein | | Descriptor: | SAKe6BE | | Authors: | Wouters, S.M.L, Noguchi, H, Velpula, G, Clarke, D.E, Voet, A.R.D, De Feyter, S. | | Deposit date: | 2021-05-25 | | Release date: | 2022-12-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | SAKe: Computationally Designed Modular Protein Building Blocks for Macromolecular Assemblies
To be published
|
|
7ONC
 
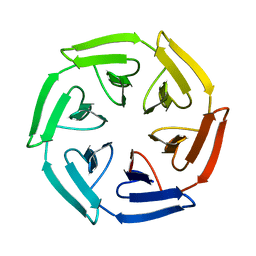 | | Crystal structure of the computationally designed SAKe6BE protein | | Descriptor: | SAKe6BE | | Authors: | Wouters, S.M.L, Noguchi, H, Velpula, G, Clarke, D.E, Voet, A.R.D, De Feyter, S. | | Deposit date: | 2021-05-25 | | Release date: | 2022-12-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | SAKe: Computationally Designed Modular Protein Building Blocks for Macromolecular Assemblies
To be published
|
|
7NBH
 
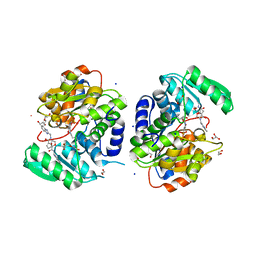 | | Crystal structure of human serine racemase in complex with DSiP fragment Z26781964, XChem fragment screen. | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Koulouris, C.R, Roe, S.M. | | Deposit date: | 2021-01-26 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Tyrosine 121 moves revealing a ligandable pocket that couples catalysis to ATP-binding in serine racemase.
Commun Biol, 5, 2022
|
|
7V8P
 
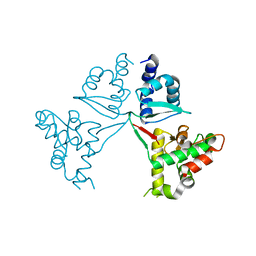 | | Crystal Structure of the MukE dimer | | Descriptor: | Chromosome partition protein MukE | | Authors: | Qian, J.W, Guo, L. | | Deposit date: | 2021-08-23 | | Release date: | 2022-01-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Crystal structure of the chromosome partition protein MukE homodimer.
Biochem.Biophys.Res.Commun., 589, 2021
|
|
7ONH
 
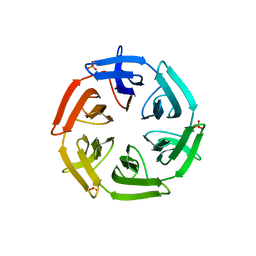 | | Crystal structure of the computationally designed SAKe6BE-L3 protein | | Descriptor: | SAKe6BE-L3, SULFATE ION | | Authors: | Wouters, S.M.L, Noguchi, H, Velpula, G, Clarke, D.E, Voet, A.R.D, De Feyter, S. | | Deposit date: | 2021-05-25 | | Release date: | 2022-12-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | SAKe: Computationally Designed Modular Protein Building Blocks for Macromolecular Assemblies
To be published
|
|
8KH3
 
 | |
5UDE
 
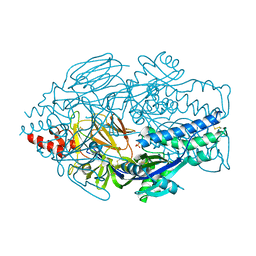 | | Crystal Structure of RSV F B9320 DS-Cav1 | | Descriptor: | Fusion glycoprotein F0, SULFATE ION | | Authors: | McLellan, J.S. | | Deposit date: | 2016-12-26 | | Release date: | 2017-05-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | A highly potent extended half-life antibody as a potential RSV vaccine surrogate for all infants.
Sci Transl Med, 9, 2017
|
|
5TZI
 
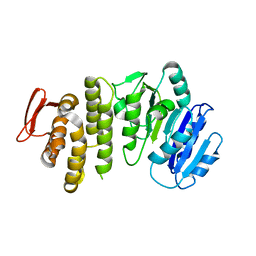 | | Crystal structure of S. aureus TarS 1-349 | | Descriptor: | Glycosyl transferase | | Authors: | Worrall, L.J, Sobhanifar, S, King, D.T, Strynadka, N.C. | | Deposit date: | 2016-11-21 | | Release date: | 2017-01-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and Mechanism of Staphylococcus aureus TarS, the Wall Teichoic Acid beta-glycosyltransferase Involved in Methicillin Resistance.
PLoS Pathog., 12, 2016
|
|
8RVB
 
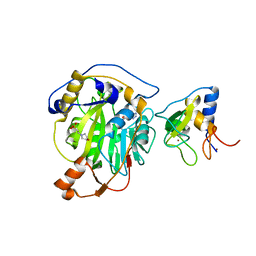 | | SARS-CoV-2 nsp16-nsp10 in complex with SAM derivative inhibitor 8 | | Descriptor: | (2~{R},3~{R},4~{S},5~{S})-2-(6-aminopurin-9-yl)-5-[2-(1~{H}-1,2,3-triazol-4-yl)ethylsulfanylmethyl]oxolane-3,4-diol, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Kalnins, G. | | Deposit date: | 2024-01-31 | | Release date: | 2024-02-14 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for inhibition of the SARS-CoV-2 nsp16 by substrate-based dual site inhibitors.
Chemmedchem, 2024
|
|
7OP4
 
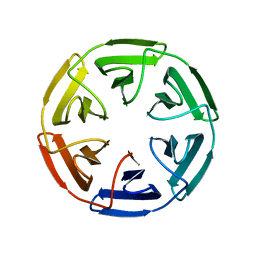 | |
7VEE
 
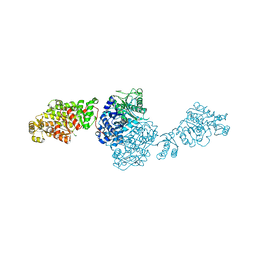 | | The ligand-free structure of GfsA KSQ-AT didomain | | Descriptor: | GLYCEROL, Polyketide synthase | | Authors: | Chisuga, T, Miyanaga, A, Nagai, A, Kudo, F, Eguchi, T. | | Deposit date: | 2021-09-08 | | Release date: | 2022-01-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Insight into the Reaction Mechanism of Ketosynthase-Like Decarboxylase in a Loading Module of Modular Polyketide Synthases.
Acs Chem.Biol., 17, 2022
|
|
7OWN
 
 | | HsNMT1 in complex with both MyrCoA and peptide AKSFSKPR | | Descriptor: | ALA-LYS-SER-PHE-SER-LYS-PRO-ARG, GLYCEROL, Glycylpeptide N-tetradecanoyltransferase 1, ... | | Authors: | Dian, C, Giglione, C, Meinnel, T. | | Deposit date: | 2021-06-18 | | Release date: | 2022-12-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Large-scale Analysis Unveil the Intertwined Paths Promoting NMT-catalyzed Lysine and Glycine Myristoylation.
J.Mol.Biol., 434, 2022
|
|
5CFR
 
 | | Crystal structure of anemone STING (Nematostella vectensis) in apo 'unrotated' closed conformation | | Descriptor: | CALCIUM ION, Stimulator of Interferon Genes | | Authors: | Kranzusch, P.J, Wilson, S.C, Lee, A.S.Y, Berger, J.M, Doudna, J.A, Vance, R.E. | | Deposit date: | 2015-07-08 | | Release date: | 2015-08-26 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Ancient Origin of cGAS-STING Reveals Mechanism of Universal 2',3' cGAMP Signaling.
Mol.Cell, 59, 2015
|
|
8UKQ
 
 | | RNA polymerase II elongation complex with Fapy-dG lesion in apo state | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Hou, P, Oh, J, Wang, D. | | Deposit date: | 2023-10-15 | | Release date: | 2024-04-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Molecular Mechanism of RNA Polymerase II Transcriptional Mutagenesis by the Epimerizable DNA Lesion, Fapy·dG.
J.Am.Chem.Soc., 146, 2024
|
|
