1VZ2
 
 | |
5N6W
 
 | | Retinoschisin R141H Mutant | | Descriptor: | Retinoschisin | | Authors: | Ramsay, E.P, Collins, R.F, Owens, T.W, Siebert, C.A, Jones, R.P.O, Roseman, A, Wang, T, Baldock, C. | | Deposit date: | 2017-02-16 | | Release date: | 2017-04-12 | | Last modified: | 2017-08-30 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural analysis of X-linked retinoschisis mutations reveals distinct classes which differentially effect retinoschisin function
Human Molecular Genetics, 25, 2016
|
|
1O6E
 
 | | Epstein-Barr virus protease | | Descriptor: | CAPSID PROTEIN P40, PHOSPHORYLISOPROPANE | | Authors: | Buisson, M, Hernandez, J, Lascoux, D, Schoehn, G, Forest, E, Arlaud, G, Seigneurin, J, Ruigrok, R.W.H, Burmeister, W.P. | | Deposit date: | 2002-09-13 | | Release date: | 2002-11-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Crystal Structure of the Epstein-Barr Virus Protease Shows Rearrangement of the Processed C Terminus
J.Mol.Biol., 324, 2002
|
|
1W1U
 
 | |
1KQC
 
 | | Structure of Nitroreductase from E. cloacae Complex with Inhibitor Acetate | | Descriptor: | ACETATE ION, FLAVIN MONONUCLEOTIDE, OXYGEN-INSENSITIVE NAD(P)H NITROREDUCTASE | | Authors: | Haynes, C.A, Koder, R.L, Miller, A.F, Rodgers, D.W. | | Deposit date: | 2002-01-04 | | Release date: | 2002-02-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of nitroreductase in three states: effects of inhibitor binding and reduction.
J.Biol.Chem., 277, 2002
|
|
1VZ3
 
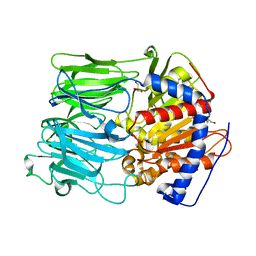 | | PROLYL OLIGOPEPTIDASE FROM PORCINE BRAIN, T597C MUTANT | | Descriptor: | GLYCEROL, PROLYL ENDOPEPTIDASE | | Authors: | Rea, D, Fulop, V. | | Deposit date: | 2004-05-14 | | Release date: | 2004-07-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Concerted Structural Changes in the Peptidase and the Propeller Domains of Prolyl Oligopeptidase are Required for Substrate Binding
J.Mol.Biol., 340, 2004
|
|
1PDN
 
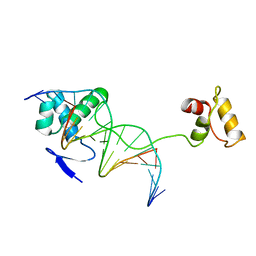 | | CRYSTAL STRUCTURE OF A PAIRED DOMAIN-DNA COMPLEX AT 2.5 ANGSTROMS RESOLUTION REVEALS STRUCTURAL BASIS FOR PAX DEVELOPMENTAL MUTATIONS | | Descriptor: | DNA (5'-D(*AP*AP*CP*GP*TP*CP*AP*CP*GP*GP*TP*TP*GP*AP*C)-3'), DNA (5'-D(*TP*TP*GP*TP*CP*AP*AP*CP*CP*GP*TP*GP*AP*CP*G)-3'), PROTEIN (PRD PAIRED) | | Authors: | Xu, W, Rould, M.A, Jun, S, Desplan, C, Pabo, C.O. | | Deposit date: | 1995-05-16 | | Release date: | 1995-07-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a paired domain-DNA complex at 2.5 A resolution reveals structural basis for Pax developmental mutations.
Cell(Cambridge,Mass.), 80, 1995
|
|
4YON
 
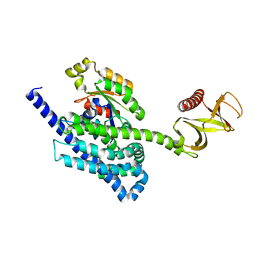 | | P-Rex1:Rac1 complex | | Descriptor: | Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 1 protein, Ras-related C3 botulinum toxin substrate 1 | | Authors: | Lucato, C.M, Whisstock, J.C, Ellisdon, A.M. | | Deposit date: | 2015-03-11 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Phosphatidylinositol (3,4,5)-Trisphosphate-dependent Rac Exchanger 1Ras-related C3 Botulinum Toxin Substrate 1 (P-Rex1Rac1) Complex Reveals the Basis of Rac1 Activation in Breast Cancer Cells.
J.Biol.Chem., 290, 2015
|
|
6B8Q
 
 | |
1KFJ
 
 | | CRYSTAL STRUCTURE OF WILD-TYPE TRYPTOPHAN SYNTHASE COMPLEXED WITH L-SERINE | | Descriptor: | SODIUM ION, TRYPTOPHAN SYNTHASE ALPHA CHAIN, TRYPTOPHAN SYNTHASE BETA CHAIN, ... | | Authors: | Kulik, V, Weyand, M, Seidel, R, Niks, D, Arac, D, Dunn, M.F, Schlichting, I. | | Deposit date: | 2001-11-21 | | Release date: | 2003-10-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | On the role of alphaThr183 in the allosteric regulation and catalytic mechanism of tryptophan synthase.
J.Mol.Biol., 324, 2002
|
|
1PDO
 
 | |
6YFV
 
 | | Crystal structure of Mtr4-Red1 minimal complex from Chaetomium thermophilum | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ATP dependent RNA helicase (Dob1)-like protein, Red1, ... | | Authors: | Dobrev, N, Ahmed, Y.L, Sinning, I. | | Deposit date: | 2020-03-26 | | Release date: | 2021-05-05 | | Last modified: | 2021-06-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The zinc-finger protein Red1 orchestrates MTREC submodules and binds the Mtl1 helicase arch domain.
Nat Commun, 12, 2021
|
|
6YGU
 
 | | Crystal structure of the minimal Mtr4-Red1 complex (single chain) from Chaetomium thermophilum | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ATP dependent RNA helicase (Dob1)-like protein, ... | | Authors: | Dobrev, N, Ahmed, Y.L, Sinning, I. | | Deposit date: | 2020-03-27 | | Release date: | 2021-05-05 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The zinc-finger protein Red1 orchestrates MTREC submodules and binds the Mtl1 helicase arch domain.
Nat Commun, 12, 2021
|
|
1YR2
 
 | |
1KFK
 
 | | Crystal structure of Tryptophan Synthase From Salmonella Typhimurium | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, TRYPTOPHAN SYNTHASE ALPHA CHAIN, ... | | Authors: | Kulik, V, Weyand, M, Seidel, R, Niks, D, Arac, D, Dunn, M.F, Schlichting, I. | | Deposit date: | 2001-11-21 | | Release date: | 2003-10-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | On the role of alphaThr183 in the allosteric regulation and catalytic mechanism of tryptophan synthase.
J.Mol.Biol., 324, 2002
|
|
1YRG
 
 | | THE CRYSTAL STRUCTURE OF RNA1P: A NEW FOLD FOR A GTPASE-ACTIVATING PROTEIN | | Descriptor: | GTPASE-ACTIVATING PROTEIN RNA1_SCHPO | | Authors: | Hillig, R.C, Renault, L, Vetter, I.R, Drell, T, Wittinghofer, A, Becker, J. | | Deposit date: | 1999-03-29 | | Release date: | 2000-03-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | The crystal structure of rna1p: a new fold for a GTPase-activating protein.
Mol.Cell, 3, 1999
|
|
1KSO
 
 | | CRYSTAL STRUCTURE OF APO S100A3 | | Descriptor: | S100 CALCIUM-BINDING PROTEIN A3 | | Authors: | Mittl, P.R, Fritz, G, Sargent, D.F, Richmond, T.J, Heizmann, C.W, Grutter, M.G. | | Deposit date: | 2002-01-14 | | Release date: | 2002-07-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Metal-free MIRAS phasing: structure of apo-S100A3.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
6B8P
 
 | |
1I54
 
 | | CYTOCHROME C (TUNA) 2FE:1ZN MIXED-METAL PORPHYRINS | | Descriptor: | CYTOCHROME C, HEME C, PROTOPORPHYRIN IX CONTAINING ZN | | Authors: | Crane, B.R, Tezcan, F.A, Winkler, J.R, Gray, H.B. | | Deposit date: | 2001-02-24 | | Release date: | 2001-05-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Electron tunneling in protein crystals.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
5XXR
 
 | |
7X08
 
 | | S protein of SARS-CoV-2 in complex with 2G1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, ... | | Authors: | Guo, Y.Y, Zhang, Y.Y, Zhou, Q. | | Deposit date: | 2022-02-21 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Broad ultra-potent neutralization of SARS-CoV-2 variants by monoclonal antibodies specific to the tip of RBD.
Cell Discov, 8, 2022
|
|
6FFA
 
 | | FMDV Leader protease bound to substrate ISG15 | | Descriptor: | GLYCEROL, Lbpro, SULFATE ION, ... | | Authors: | Swatek, K.N, Pruneda, J.N, Komander, D. | | Deposit date: | 2018-01-05 | | Release date: | 2018-02-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Irreversible inactivation of ISG15 by a viral leader protease enables alternative infection detection strategies.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4WPA
 
 | |
6PTL
 
 | |
1ID1
 
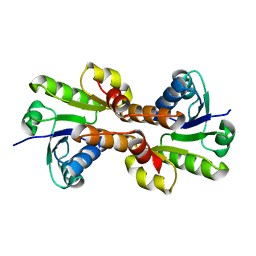 | | CRYSTAL STRUCTURE OF THE RCK DOMAIN FROM E.COLI POTASSIUM CHANNEL | | Descriptor: | PUTATIVE POTASSIUM CHANNEL PROTEIN | | Authors: | Jiang, Y, Pico, A, Cadene, M, Chait, B.T, MacKinnon, R. | | Deposit date: | 2001-04-02 | | Release date: | 2001-04-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the RCK domain from the E. coli K+ channel and demonstration of its presence in the human BK channel.
Neuron, 29, 2001
|
|
