8RVE
 
 | | Vimentin intermediate filament | | Descriptor: | Vimentin | | Authors: | Eibauer, M, Medalia, O. | | Deposit date: | 2024-02-01 | | Release date: | 2024-04-10 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Vimentin filaments integrate low-complexity domains in a complex helical structure.
Nat.Struct.Mol.Biol., 31, 2024
|
|
1BVQ
 
 | |
6VU8
 
 | | Structure of G-alpha-i bound to its chaperone Ric-8A | | Descriptor: | Guanine nucleotide-binding protein G(i) subunit alpha-1, Resistance to inhibitors of cholinesterase 8 homolog A (C. elegans) | | Authors: | Seven, A.B, Hilger, D. | | Deposit date: | 2020-02-14 | | Release date: | 2020-03-18 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.14 Å) | | Cite: | Structures of G alpha Proteins in Complex with Their Chaperone Reveal Quality Control Mechanisms.
Cell Rep, 30, 2020
|
|
1BRK
 
 | | BARNASE MUTANT WITH ILE 96 REPLACED BY ALA | | Descriptor: | BARNASE, ZINC ION | | Authors: | Cramer, P.C, Buckle, A, Fersht, A. | | Deposit date: | 1995-03-09 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and energetic responses to cavity-creating mutations in hydrophobic cores: observation of a buried water molecule and the hydrophilic nature of such hydrophobic cavities.
Biochemistry, 35, 1996
|
|
6XZ8
 
 | | Structure of aldosterone synthase (CYP11B2) in complex with N-[(1R)-1-[5-(6-chloro-1,1-dimethyl-3-oxo-isoindolin-2-yl)-3-pyridyl]ethyl]methanesulfonamide | | Descriptor: | Cytochrome P450 11B2, mitochondrial, HEME C, ... | | Authors: | Kuglstatter, A, Joseph, C, Benz, J. | | Deposit date: | 2020-02-03 | | Release date: | 2020-06-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Discovery of 3-Pyridyl Isoindolin-1-one Derivatives as Potent, Selective, and Orally Active Aldosterone Synthase (CYP11B2) Inhibitors.
J.Med.Chem., 63, 2020
|
|
7MYR
 
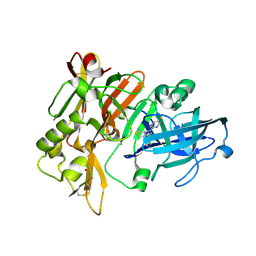 | | BACE-1 in complex with compound #18 | | Descriptor: | (4aR,7aR)-6-(5-fluoropyrimidin-2-yl)-7a-(1,2-thiazol-5-yl)-4,4a,5,6,7,7a-hexahydropyrrolo[3,4-d][1,3]thiazin-2-amine, Beta-secretase 1, GLYCEROL, ... | | Authors: | Hendle, J, Timm, D.E, Stout, S.L. | | Deposit date: | 2021-05-21 | | Release date: | 2021-07-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Discovery and Early Clinical Development of LY3202626, a Low-Dose, CNS-Penetrant BACE Inhibitor.
J.Med.Chem., 64, 2021
|
|
7MYU
 
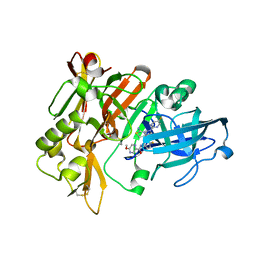 | | BACE-1 in complex with compound #22 | | Descriptor: | Beta-secretase 1, N-{3-[(4aR,7aS)-2-amino-6-(5-fluoropyrimidin-2-yl)-4a,5,6,7-tetrahydropyrrolo[3,4-d][1,3]thiazin-7a(4H)-yl]-4-fluorophenyl}-5-methoxypyrazine-2-carboxamide, SULFATE ION | | Authors: | Hendle, J, Timm, D.E, Stout, S.L. | | Deposit date: | 2021-05-21 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Discovery and Early Clinical Development of LY3202626, a Low-Dose, CNS-Penetrant BACE Inhibitor.
J.Med.Chem., 64, 2021
|
|
6XBJ
 
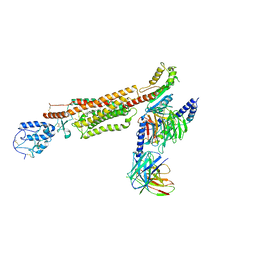 | | Structure of human SMO-D384R complex with Gi | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Qi, X, Long, T, Li, X. | | Deposit date: | 2020-06-06 | | Release date: | 2020-09-30 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | Sterols in an intramolecular channel of Smoothened mediate Hedgehog signaling.
Nat.Chem.Biol., 16, 2020
|
|
7MYI
 
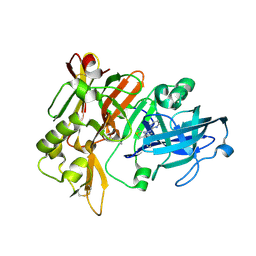 | | BACE-1 in complex with compound #6 | | Descriptor: | (4aR,7aR)-6-(pyrimidin-2-yl)-7a-(thiophen-2-yl)-4,4a,5,6,7,7a-hexahydropyrrolo[3,4-d][1,3]thiazin-2-amine, Beta-secretase 1, GLYCEROL | | Authors: | Hendle, J, Timm, D.E, Stout, S.L. | | Deposit date: | 2021-05-21 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Discovery and Early Clinical Development of LY3202626, a Low-Dose, CNS-Penetrant BACE Inhibitor.
J.Med.Chem., 64, 2021
|
|
1BLR
 
 | | NMR SOLUTION STRUCTURE OF HUMAN CELLULAR RETINOIC ACID BINDING PROTEIN-TYPE II, 22 STRUCTURES | | Descriptor: | CELLULAR RETINOIC ACID BINDING PROTEIN-TYPE II | | Authors: | Wang, L, Li, Y, Abilddard, F, Yan, H, Markely, J. | | Deposit date: | 1998-07-20 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of type II human cellular retinoic acid binding protein: implications for ligand binding.
Biochemistry, 37, 1998
|
|
1OBH
 
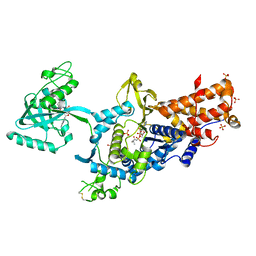 | | LEUCYL-TRNA SYNTHETASE FROM THERMUS THERMOPHILUS COMPLEXED WITH A PRE-TRANSFER EDITING SUBSTRATE ANALOGUE IN BOTH SYNTHETIC ACTIVE SITE AND EDITING SITE | | Descriptor: | LEUCYL-TRNA SYNTHETASE, MERCURY (II) ION, NORVALINE, ... | | Authors: | Cusack, S, Yaremchuk, A, Tukalo, M. | | Deposit date: | 2003-01-31 | | Release date: | 2003-05-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Mechanistic Basis of Pre- and Posttransfer Editing by Leucyl-tRNA Synthetase
Mol.Cell, 11, 2003
|
|
1O66
 
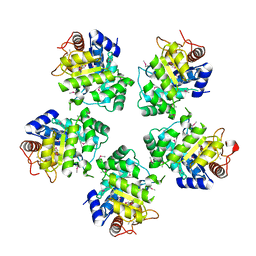 | |
2IGO
 
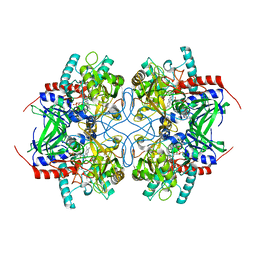 | |
6XVQ
 
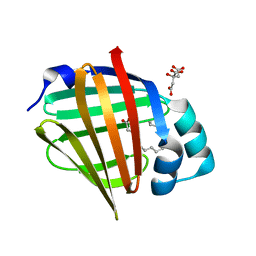 | | Human myelin protein P2 mutant K31Q | | Descriptor: | CITRIC ACID, Myelin P2 protein, PALMITIC ACID | | Authors: | Ruskamo, S, Lehtimaki, M, Kursula, P. | | Deposit date: | 2020-01-22 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cryo-EM, X-ray diffraction, and atomistic simulations reveal determinants for the formation of a supramolecular myelin-like proteolipid lattice.
J.Biol.Chem., 295, 2020
|
|
6XVY
 
 | | Human myelin protein P2 mutant R88Q | | Descriptor: | CHLORIDE ION, CITRIC ACID, Myelin P2 protein, ... | | Authors: | Ruskamo, S, Lehtimaki, M, Kursula, P. | | Deposit date: | 2020-01-22 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cryo-EM, X-ray diffraction, and atomistic simulations reveal determinants for the formation of a supramolecular myelin-like proteolipid lattice.
J.Biol.Chem., 295, 2020
|
|
7NSL
 
 | |
7O89
 
 | | sulerythrin without metals (apo-state) | | Descriptor: | CHLORIDE ION, Sulerythrin | | Authors: | Jeoung, J.-H, Dobbek, H. | | Deposit date: | 2021-04-15 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Bimetallic Mn, Fe, Co, and Ni Sites in a Four-Helix Bundle Protein: Metal Binding, Structure, and Peroxide Activation.
Inorg.Chem., 60, 2021
|
|
7O99
 
 | | diMn-sulerythrin | | Descriptor: | CHLORIDE ION, COBALT (II) ION, Sulerythrin | | Authors: | Jeoung, J.-H, Dobbek, H. | | Deposit date: | 2021-04-15 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Bimetallic Mn, Fe, Co, and Ni Sites in a Four-Helix Bundle Protein: Metal Binding, Structure, and Peroxide Activation.
Inorg.Chem., 60, 2021
|
|
7O90
 
 | | Mono-Fe-sulerythrin | | Descriptor: | CHLORIDE ION, FE (III) ION, Sulerythrin | | Authors: | Jeoung, J.-H, Dobbek, H. | | Deposit date: | 2021-04-15 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Bimetallic Mn, Fe, Co, and Ni Sites in a Four-Helix Bundle Protein: Metal Binding, Structure, and Peroxide Activation.
Inorg.Chem., 60, 2021
|
|
7O93
 
 | | diMn-sulerythrin | | Descriptor: | (R,R)-2,3-BUTANEDIOL, CHLORIDE ION, HYDROGEN PEROXIDE, ... | | Authors: | Jeoung, J.-H, Dobbek, H. | | Deposit date: | 2021-04-15 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Bimetallic Mn, Fe, Co, and Ni Sites in a Four-Helix Bundle Protein: Metal Binding, Structure, and Peroxide Activation.
Inorg.Chem., 60, 2021
|
|
7O91
 
 | | diMn-sulerythrin | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, MANGANESE (II) ION, ... | | Authors: | Jeoung, J.-H, Dobbek, H. | | Deposit date: | 2021-04-15 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Bimetallic Mn, Fe, Co, and Ni Sites in a Four-Helix Bundle Protein: Metal Binding, Structure, and Peroxide Activation.
Inorg.Chem., 60, 2021
|
|
7O9C
 
 | | diNi-sulerythrin treated by hydrogen perxoide | | Descriptor: | CHLORIDE ION, NICKEL (II) ION, Sulerythrin | | Authors: | Jeoung, J.-H, Dobbek, H. | | Deposit date: | 2021-04-15 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Bimetallic Mn, Fe, Co, and Ni Sites in a Four-Helix Bundle Protein: Metal Binding, Structure, and Peroxide Activation.
Inorg.Chem., 60, 2021
|
|
7O9E
 
 | | diNi-sulerythrin | | Descriptor: | CHLORIDE ION, NICKEL (II) ION, Sulerythrin | | Authors: | Jeoung, J.-H, Dobbek, H. | | Deposit date: | 2021-04-15 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Bimetallic Mn, Fe, Co, and Ni Sites in a Four-Helix Bundle Protein: Metal Binding, Structure, and Peroxide Activation.
Inorg.Chem., 60, 2021
|
|
7O8D
 
 | | diFe-sulerythrin oxidised by H2O2 | | Descriptor: | CHLORIDE ION, FE (III) ION, Sulerythrin | | Authors: | Jeoung, J.-H, Dobbek, H. | | Deposit date: | 2021-04-15 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Bimetallic Mn, Fe, Co, and Ni Sites in a Four-Helix Bundle Protein: Metal Binding, Structure, and Peroxide Activation.
Inorg.Chem., 60, 2021
|
|
7O9D
 
 | | peroxide-bound diCo-sulerythrin | | Descriptor: | CHLORIDE ION, COBALT (II) ION, HYDROGEN PEROXIDE, ... | | Authors: | Jeoung, J.-H, Dobbek, H. | | Deposit date: | 2021-04-15 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Bimetallic Mn, Fe, Co, and Ni Sites in a Four-Helix Bundle Protein: Metal Binding, Structure, and Peroxide Activation.
Inorg.Chem., 60, 2021
|
|
