3ZQX
 
 | | Carbohydrate-binding module CBM3b from the cellulosomal cellobiohydrolase 9A from Clostridium thermocellum | | Descriptor: | CALCIUM ION, CELLULOSE 1,4-BETA-CELLOBIOSIDASE | | Authors: | Yaniv, O, Petkun, S, Shimon, L.J.W, Bayer, E.A, Lamed, R, Frolow, F. | | Deposit date: | 2011-06-12 | | Release date: | 2012-04-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | A Single Mutation Reforms the Binding Activity of an Adhesion-Deficient Family 3 Carbohydrate-Binding Module
Acta Crystallogr.,Sect.D, 68, 2012
|
|
2GZR
 
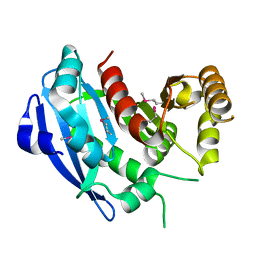 | |
2D2F
 
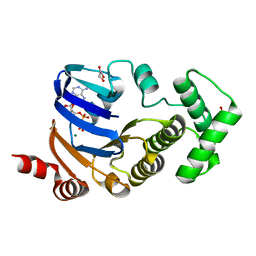 | | Crystal structure of atypical cytoplasmic ABC-ATPase SufC from Thermus thermophilus HB8 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Watanabe, S, Kita, A, Miki, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-09-08 | | Release date: | 2005-10-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Atypical Cytoplasmic ABC-ATPase SufC from Thermus thermophilus HB8.
J.Mol.Biol., 353, 2005
|
|
2GJ5
 
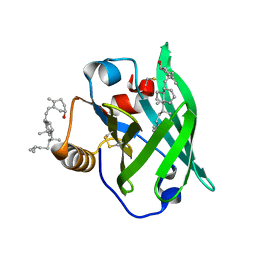 | | Crystal structure of a secondary vitamin D3 binding site of milk beta-lactoglobulin | | Descriptor: | (1S,3Z)-3-[(2E)-2-[(1R,3AR,7AS)-7A-METHYL-1-[(2R)-6-METHYLHEPTAN-2-YL]-2,3,3A,5,6,7-HEXAHYDRO-1H-INDEN-4-YLIDENE]ETHYLI DENE]-4-METHYLIDENE-CYCLOHEXAN-1-OL, Beta-lactoglobulin | | Authors: | Yang, M.C, Guan, H.H, Liu, M.Y, Yang, J.M, Chen, W.L, Chen, C.J, Mao, S.J. | | Deposit date: | 2006-03-30 | | Release date: | 2007-10-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a secondary vitamin D3 binding site of milk beta-lactoglobulin.
Proteins, 71, 2008
|
|
2GZS
 
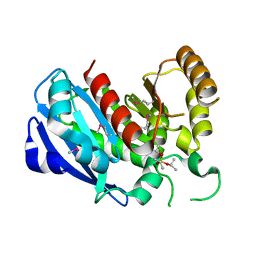 | |
2ANE
 
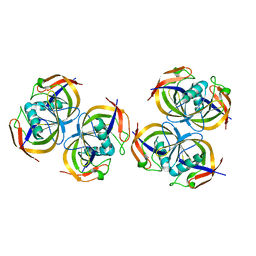 | | Crystal structure of N-terminal domain of E.Coli Lon Protease | | Descriptor: | ATP-dependent protease La | | Authors: | Li, M, Rasulova, F, Melnikov, E.E, Rotanova, T.V, Gustchina, A, Maurizi, M.R, Wlodawer, A. | | Deposit date: | 2005-08-11 | | Release date: | 2005-11-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structure of the N-terminal domain of E. coli Lon protease.
Protein Sci., 14, 2005
|
|
4BNQ
 
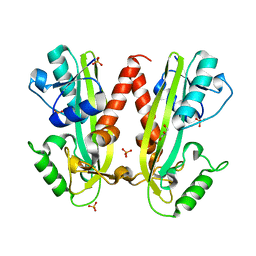 | |
2M5E
 
 | | Structure of the C-domain of Calcium-saturated Calmodulin bound to the IQ motif of NaV1.2 | | Descriptor: | CALCIUM ION, Calmodulin, Sodium channel protein type 2 subunit alpha | | Authors: | Fowler, C.A, Feldkamp, M.D, Yu, L, Shea, M.A. | | Deposit date: | 2013-02-21 | | Release date: | 2014-07-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Calcium triggers reversal of calmodulin on nested anti-parallel sites in the IQ motif of the neuronal voltage-dependent sodium channel NaV1.2.
Biophys. Chem., 224, 2017
|
|
4CFY
 
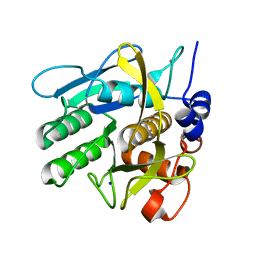 | | SAVINASE CRYSTAL STRUCTURES FOR COMBINED SINGLE CRYSTAL DIFFRACTION AND POWDER DIFFRACTION ANALYSIS | | Descriptor: | CALCIUM ION, SODIUM ION, SUBTILISIN SAVINASE | | Authors: | Frankaer, C.G, Moroz, O.V, Turkenburg, J.P, Aspmo, S.I, Thymark, M, Friis, E.P, Stahla, K, Nielsen, J.E, Wilson, K.S, Harris, P. | | Deposit date: | 2013-11-19 | | Release date: | 2014-04-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Analysis of an Industrial Production Suspension of Bacillus Lentus Subtilisin Crystals by Powder Diffraction: A Powerful Quality-Control Tool.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2MUH
 
 | |
2N1O
 
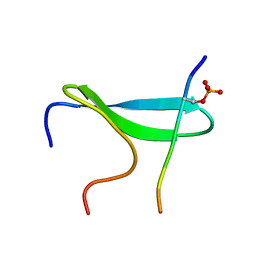 | |
2F8B
 
 | |
2K5X
 
 | |
3UJP
 
 | | Structure of MntC protein at 2.7A | | Descriptor: | CACODYLATE ION, MANGANESE (II) ION, Mn transporter subunit, ... | | Authors: | Kanteev, M, Adir, N. | | Deposit date: | 2011-11-08 | | Release date: | 2012-11-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Arginine 116 stabilizes the entrance to the metal ion-binding site of the MntC protein.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
2FKX
 
 | |
2I9S
 
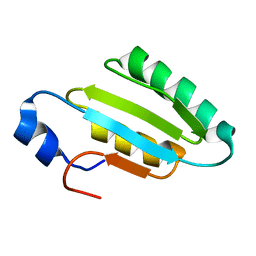 | | The solution structure of the core of mesoderm development (MESD). | | Descriptor: | Mesoderm development candidate 2 | | Authors: | Koehler, C, Andersen, O, Diehl, A, Schmieder, P, Krause, G, Oschkinat, H. | | Deposit date: | 2006-09-06 | | Release date: | 2007-05-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the core of mesoderm development (MESD), a chaperone for members of the LDLR-family
J.STRUCT.FUNCT.GENOM., 7, 2006
|
|
2MEM
 
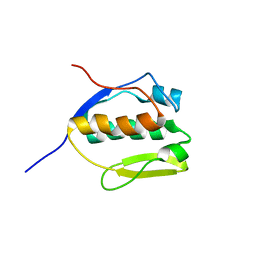 | | Solution NMR structure of SLED domain of Scml2 | | Descriptor: | Sex comb on midleg-like protein 2 | | Authors: | Bezsonova, I. | | Deposit date: | 2013-09-24 | | Release date: | 2014-04-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of the DNA-binding Domain from Scml2 (Sex Comb on Midleg-like 2).
J.Biol.Chem., 289, 2014
|
|
2LXZ
 
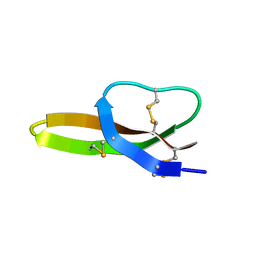 | | Solution Structure of the Antimicrobial Peptide Human Defensin 5 | | Descriptor: | Defensin-5 | | Authors: | Wommack, A.J, Robson, S.A, Wanniarahchi, Y.A, Wan, A, Turner, C.J, Nolan, E.M. | | Deposit date: | 2012-09-10 | | Release date: | 2012-11-28 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure and condition-dependent oligomerization of the antimicrobial Peptide human defensin 5.
Biochemistry, 51, 2012
|
|
2OQY
 
 | | The crystal structure of muconate cycloisomerase from Oceanobacillus iheyensis | | Descriptor: | MAGNESIUM ION, Muconate cycloisomerase | | Authors: | Fedorov, A.A, Toro, R, Fedorov, E.V, Bonanno, J, Sauder, J.M, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-02-01 | | Release date: | 2007-03-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Computation-facilitated assignment of the function in the enolase superfamily: a regiochemically distinct galactarate dehydratase from Oceanobacillus iheyensis .
Biochemistry, 48, 2009
|
|
3VCY
 
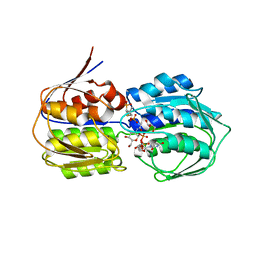 | | Structure of MurA (UDP-N-acetylglucosamine enolpyruvyl transferase), from Vibrio fischeri in complex with substrate UDP-N-acetylglucosamine and the drug fosfomycin. | | Descriptor: | GLYCEROL, PHOSPHATE ION, UDP-N-acetylglucosamine 1-carboxyvinyltransferase, ... | | Authors: | Bensen, D.C, Rodriguez, S, Nix, J, Cunningham, M.L, Tari, L.W. | | Deposit date: | 2012-01-04 | | Release date: | 2012-04-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.925 Å) | | Cite: | Structure of MurA (UDP-N-acetylglucosamine enolpyruvyl transferase) from Vibrio fischeri in complex with substrate UDP-N-acetylglucosamine and the drug fosfomycin.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
2KVJ
 
 | | NMR and MD solution structure of a Gamma-Methylated PNA duplex | | Descriptor: | Gamma-Modified Peptide Nucleic Acid | | Authors: | He, W, Crawford, M.J, Rapireddy, S, Madrid, M, Gil, R.R, Ly, D.H, Achim, C. | | Deposit date: | 2010-03-15 | | Release date: | 2010-05-12 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | The structure of a gamma-modified peptide nucleic acid duplex.
Mol Biosyst, 6, 2010
|
|
2EIO
 
 | |
2EIQ
 
 | |
2M3M
 
 | |
3U1R
 
 | | Structure Analysis of A New Psychrophilic Marine Protease | | Descriptor: | Alkaline metalloprotease, CALCIUM ION, ZINC ION | | Authors: | Zhang, S.-C, Sun, M, Tang, L, Wang, Q.-H, Hao, J.-H, Han, Y, Hu, X.-J, Zhou, M, Lin, S.-X. | | Deposit date: | 2011-09-30 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure analysis of a new psychrophilic marine protease.
Plos One, 6, 2011
|
|
