1C6F
 
 | |
1C6P
 
 | |
1CCQ
 
 | |
7BTF
 
 | | SARS-CoV-2 RNA-dependent RNA polymerase in complex with cofactors in reduced condition | | Descriptor: | Non-structural protein 7, Non-structural protein 8, RNA-directed RNA polymerase, ... | | Authors: | Gao, Y, Yan, L, Huang, Y, Liu, F, Cao, L, Wang, T, Wang, Q, Lou, Z, Rao, Z. | | Deposit date: | 2020-04-01 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structure of the RNA-dependent RNA polymerase from COVID-19 virus.
Science, 368, 2020
|
|
1CDW
 
 | | HUMAN TBP CORE DOMAIN COMPLEXED WITH DNA | | Descriptor: | DNA (5'-D(*CP*AP*GP*CP*CP*TP*TP*TP*TP*AP*TP*AP*GP*CP*AP*G)-3'), DNA (5'-D(*CP*TP*GP*CP*TP*AP*TP*AP*AP*AP*AP*GP*GP*CP*TP*G)-3'), PROTEIN (TATA BINDING PROTEIN (TBP)) | | Authors: | Nikolov, D.B, Chen, H, Halay, E.D, Hoffmann, A, Roeder, R.G, Burley, S.K. | | Deposit date: | 1996-04-11 | | Release date: | 1996-12-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a human TATA box-binding protein/TATA element complex.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
1C6H
 
 | |
1C6R
 
 | | CRYSTAL STRUCTURE OF REDUCED CYTOCHROME C6 FROM THE GREEN ALGAE SCENEDESMUS OBLIQUUS | | Descriptor: | CADMIUM ION, CYTOCHROME C6, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Schnackenberg, J, Than, M.E, Mann, K, Wiegand, G, Huber, R, Reuter, W. | | Deposit date: | 1999-04-06 | | Release date: | 2000-04-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Amino acid sequence, crystallization and structure determination of reduced and oxidized cytochrome c6 from the green alga Scenedesmus obliquus.
J.Mol.Biol., 290, 1999
|
|
1C6G
 
 | |
1C6Q
 
 | |
1C8X
 
 | | Endo-Beta-N-Acetylglucosaminidase H, D130E Mutant | | Descriptor: | ENDO-BETA-N-ACETYLGLUCOSAMINIDASE H, PHOSPHATE ION | | Authors: | Rao, V, Tao, C, Guan, C, Van Roey, P. | | Deposit date: | 1999-07-30 | | Release date: | 1999-11-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutations of endo-beta-N-acetylglucosaminidase H active site residues Asp130 and Glu132: activities and conformations.
Protein Sci., 8, 1999
|
|
1C8I
 
 | | BINDING MODE OF HYDROXYLAMINE TO ARTHROMYCES RAMOSUS PEROXIDASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, HYDROXYAMINE, ... | | Authors: | Wariishi, H, Nonaka, D, Johjima, T, Nakamura, N, Naruta, Y, Kubo, K, Fukuyama, K. | | Deposit date: | 2000-05-08 | | Release date: | 2001-01-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Direct binding of hydroxylamine to the heme iron of Arthromyces ramosus peroxidase. Substrate analogue that inhibits compound I formation in a competetive manner.
J.Biol.Chem., 275, 2000
|
|
1C91
 
 | | Endo-Beta-N-Acetylglucosaminidase H, E132D | | Descriptor: | ENDO-BETA-N-ACETYLGLUCOSAMINIDASE H | | Authors: | Rao, V, Cui, T, Guan, C, Van Roey, P. | | Deposit date: | 1999-07-30 | | Release date: | 1999-11-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mutations of endo-beta-N-acetylglucosaminidase H active site residues Asp130 and Glu132: activities and conformations.
Protein Sci., 8, 1999
|
|
1CCB
 
 | | THE ASP-HIS-FE TRIAD OF CYTOCHROME C PEROXIDASE CONTROLS THE REDUCTION POTENTIAL, ELECTRONIC STRUCTURE, AND COUPLING OF THE TRYPTOPHAN FREE-RADICAL TO THE HEME | | Descriptor: | CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Goodin, D.B, Mcree, D.E. | | Deposit date: | 1993-01-04 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Asp-His-Fe triad of cytochrome c peroxidase controls the reduction potential, electronic structure, and coupling of the tryptophan free radical to the heme.
Biochemistry, 32, 1993
|
|
7F9X
 
 | | Structure of the second OTU domain of LotA | | Descriptor: | LotA | | Authors: | Iwai, T, Takekawa, T, Kubori, T, Nagai, H, Imada, K. | | Deposit date: | 2021-07-05 | | Release date: | 2021-10-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural Basis of Ubiquitin Recognition by a Bacterial Ovarian Tumor Deubiquitinase LotA.
J.Bacteriol., 204, 2022
|
|
1CDM
 
 | |
1C8Y
 
 | | Endo-Beta-N-Acetylglucosaminidase H, D130A Mutant | | Descriptor: | ENDO-BETA-N-ACETYLGLUCOSAMINIDASE H, ZINC ION | | Authors: | Rao, V, Cui, T, Guan, C, Van Roey, P. | | Deposit date: | 1999-07-30 | | Release date: | 1999-11-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutations of endo-beta-N-acetylglucosaminidase H active site residues Asp130 and Glu132: activities and conformations.
Protein Sci., 8, 1999
|
|
7BCM
 
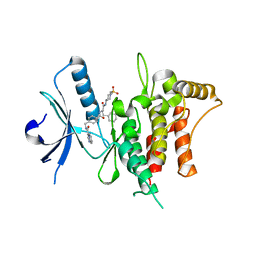 | | The DDR1 Kinase Domain Bound To SR302 | | Descriptor: | Epithelial discoidin domain-containing receptor 1, ~{N}-[[4-[[(2~{S})-4-cyclohexyl-1-[[(3~{S})-1-methylsulfonylpiperidin-3-yl]amino]-1-oxidanylidene-butan-2-yl]carbamoyl]phenyl]methyl]imidazo[1,2-a]pyridine-3-carboxamide | | Authors: | Mathea, S, Chatterjee, D, Preuss, F, Roehm, S, Joerger, A, Knapp, S. | | Deposit date: | 2020-12-20 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Development of a Selective Dual Discoidin Domain Receptor (DDR)/p38 Kinase Chemical Probe.
J.Med.Chem., 64, 2021
|
|
1C9B
 
 | |
8TAZ
 
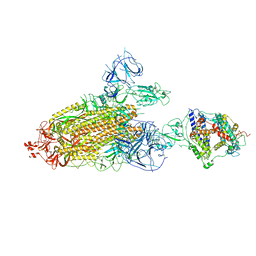 | | Cryo-EM structure of mink variant Y453F trimeric spike protein bound to one mink ACE2 receptors | | Descriptor: | Angiotensin-converting enzyme, Spike glycoprotein | | Authors: | Ahn, H.M, Calderon, B, Fan, X, Gao, Y, Horgan, N, Liang, B. | | Deposit date: | 2023-06-28 | | Release date: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Structural basis of the American mink ACE2 binding by Y453F trimeric spike glycoproteins of SARS-CoV-2.
J Med Virol, 95, 2023
|
|
1C6I
 
 | |
1C6T
 
 | |
1CCC
 
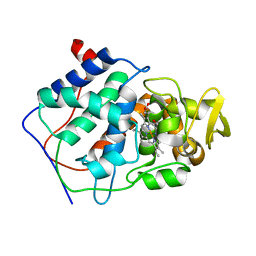 | | THE ASP-HIS-FE TRIAD OF CYTOCHROME C PEROXIDASE CONTROLS THE REDUCTION POTENTIAL, ELECTRONIC STRUCTURE, AND COUPLING OF THE TRYPTOPHAN FREE-RADICAL TO THE HEME | | Descriptor: | CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Goodin, D.B, Mcree, D.E. | | Deposit date: | 1993-01-04 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Asp-His-Fe triad of cytochrome c peroxidase controls the reduction potential, electronic structure, and coupling of the tryptophan free radical to the heme.
Biochemistry, 32, 1993
|
|
8T21
 
 | | Cryo-EM structure of mink variant Y453F trimeric spike protein | | Descriptor: | Spike glycoprotein | | Authors: | Ahn, H.M, Calderon, B, Fan, X, Gao, Y, Horgan, N, Zhou, B, Liang, B. | | Deposit date: | 2023-06-05 | | Release date: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of the American mink ACE2 binding by Y453F trimeric spike glycoproteins of SARS-CoV-2.
J Med Virol, 95, 2023
|
|
7BEP
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in a ternary complex with COVOX-384 and S309 Fabs | | Descriptor: | CHLORIDE ION, COVOX-384 heavy chain, COVOX-384 light chain, ... | | Authors: | Zhou, D, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2020-12-24 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
8T22
 
 | | Cryo-EM structure of mink variant Y453F trimeric spike protein bound to one mink ACE2 receptors at downRBD conformation | | Descriptor: | Angiotensin-converting enzyme, Spike glycoprotein | | Authors: | Ahn, H.M, Calderon, B, Fan, X, Gao, Y, Horgan, N, Zhou, B, Liang, B. | | Deposit date: | 2023-06-05 | | Release date: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.83 Å) | | Cite: | Structural basis of the American mink ACE2 binding by Y453F trimeric spike glycoproteins of SARS-CoV-2.
J Med Virol, 95, 2023
|
|
