8QXM
 
 | | Cryo-EM structure of tetrameric human SAMHD1 State III - Relaxed | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, ... | | Authors: | Acton, O.J, Sheppard, D, Rosenthal, P.B, Taylor, I.A. | | Deposit date: | 2023-10-24 | | Release date: | 2024-05-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Platform-directed allostery and quaternary structure dynamics of SAMHD1 catalysis.
Nat Commun, 15, 2024
|
|
8RHQ
 
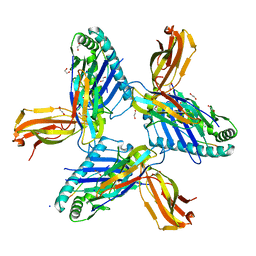 | | Crystal structure of HLA-A*11:01 in complex with SVLNDIFSRL, an 10-mer epitope from SARS-CoV-2 Spike (S975-984) | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, CHLORIDE ION, ... | | Authors: | Ahn, Y.M, Maddumage, J.C, Szeto, C, Gras, S. | | Deposit date: | 2023-12-16 | | Release date: | 2024-05-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The impact of SARS-CoV-2 spike mutation on peptide presentation is HLA allomorph-specific.
Curr Res Struct Biol, 7, 2024
|
|
8BZ5
 
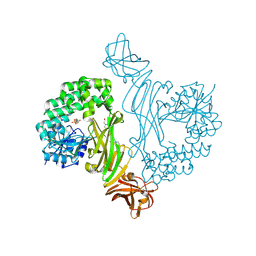 | |
8BZ8
 
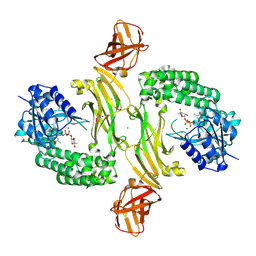 | |
7RDY
 
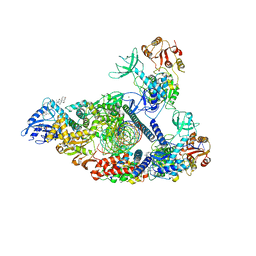 | | SARS-CoV-2 replication-transcription complex bound to nsp13 helicase - nsp13(2)-RTC - engaged class | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, CHAPSO, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2021-07-12 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Ensemble cryo-EM reveals conformational states of the nsp13 helicase in the SARS-CoV-2 helicase replication-transcription complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RE0
 
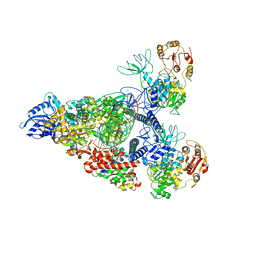 | | SARS-CoV-2 replication-transcription complex bound to nsp13 helicase - nsp13(2)-RTC - swiveled class | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Helicase, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2021-07-12 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Ensemble cryo-EM reveals conformational states of the nsp13 helicase in the SARS-CoV-2 helicase replication-transcription complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
5IPE
 
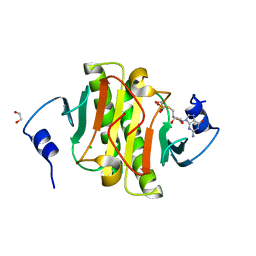 | | Human Histidine Triad Nucleotide Binding Protein 1 (hHint1) nucleoside thiophosphoramidate catalytic product complex | | Descriptor: | 1,2-ETHANEDIOL, 5'-S-phosphono-5'-thioguanosine, CHLORIDE ION, ... | | Authors: | Maize, K.M, Finzel, B.C. | | Deposit date: | 2016-03-09 | | Release date: | 2017-03-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Caught before Released: Structural Mapping of the Reaction Trajectory for the Sofosbuvir Activating Enzyme, Human Histidine Triad Nucleotide Binding Protein 1 (hHint1).
Biochemistry, 56, 2017
|
|
8QXJ
 
 | | Cryo-EM structure of tetrameric human SAMHD1 with dApNHpp | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, FE (III) ION, ... | | Authors: | Acton, O.J, Sheppard, D, Rosenthal, P.B, Taylor, I.A. | | Deposit date: | 2023-10-24 | | Release date: | 2024-05-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Platform-directed allostery and quaternary structure dynamics of SAMHD1 catalysis.
Nat Commun, 15, 2024
|
|
5MDO
 
 | |
7RDS
 
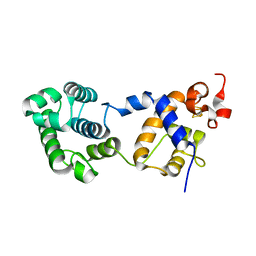 | | Structure of human NTHL1 | | Descriptor: | IRON/SULFUR CLUSTER, Isoform 3 of Endonuclease III-like protein 1 | | Authors: | Carroll, B.L, Zahn, K.E, Doublie, S. | | Deposit date: | 2021-07-11 | | Release date: | 2021-12-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Caught in motion: human NTHL1 undergoes interdomain rearrangement necessary for catalysis.
Nucleic Acids Res., 49, 2021
|
|
1CB8
 
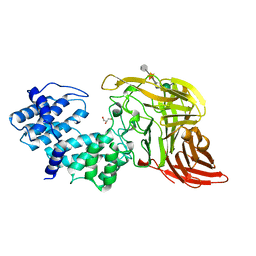 | | CHONDROITINASE AC LYASE FROM FLAVOBACTERIUM HEPARINUM | | Descriptor: | CALCIUM ION, GLYCEROL, PROTEIN (CHONDROITINASE AC), ... | | Authors: | Fethiere, J, Eggimann, B, Cygler, M. | | Deposit date: | 1999-03-02 | | Release date: | 1999-05-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of chondroitin AC lyase, a representative of a family of glycosaminoglycan degrading enzymes.
J.Mol.Biol., 288, 1999
|
|
6U1L
 
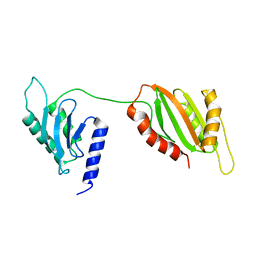 | | Structure of two-domain translational regulator Yih1 reveals a possible mechanism of action | | Descriptor: | Protein IMPACT homolog | | Authors: | Harjes, E, Jameson, G.B, Edwards, P.J.B, Goroncy, A.K, Loo, T, Norris, G.E. | | Deposit date: | 2019-08-16 | | Release date: | 2021-02-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Experimentally based structural model of Yih1 provides insight into its function in controlling the key translational regulator Gcn2.
Febs Lett., 595, 2021
|
|
6U1O
 
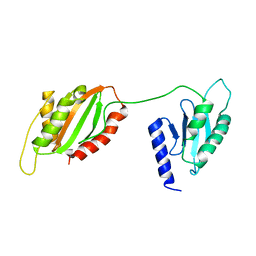 | | Structure of two-domain translational regulator Yih1 reveals a possible mechanism of action | | Descriptor: | Protein IMPACT homolog | | Authors: | Harjes, E, Jameson, G.B, Edwards, P.J.B, Goroncy, A.K, Loo, T, Norris, G.E. | | Deposit date: | 2019-08-16 | | Release date: | 2021-02-10 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Experimentally based structural model of Yih1 provides insight into its function in controlling the key translational regulator Gcn2.
Febs Lett., 595, 2021
|
|
7RJZ
 
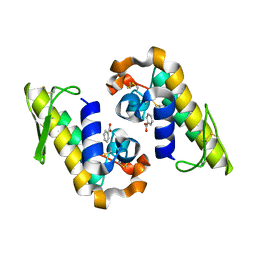 | |
6Z5X
 
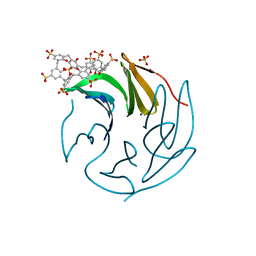 | | The RSL - sulfonato-calix[8]arene complex, P213 form, acetate pH 4.8 | | Descriptor: | Fucose-binding lectin protein, SODIUM ION, SULFATE ION, ... | | Authors: | Engilberge, S, Ramberg, K, Crowley, P.B. | | Deposit date: | 2020-05-27 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Facile Fabrication of Protein-Macrocycle Frameworks.
J.Am.Chem.Soc., 143, 2021
|
|
7ZMW
 
 | |
6NUK
 
 | | De novo designed protein Ferredog-Diesel | | Descriptor: | Ferredog-Diesel | | Authors: | Koepnick, B, Bick, M.J, DiMaio, F, Norgard-Solano, T, Baker, D. | | Deposit date: | 2019-02-01 | | Release date: | 2019-06-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | De novo protein design by citizen scientists.
Nature, 570, 2019
|
|
6Q5G
 
 | |
6Z5G
 
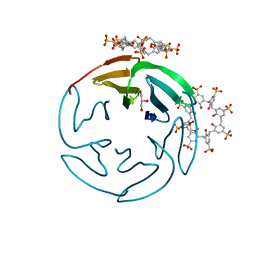 | | The RSL - sulfonato-calix[8]arene complex, I23 form, citrate pH 4.0, solved by S-SAD | | Descriptor: | Fucose-binding lectin protein, GLYCEROL, beta-D-fructopyranose, ... | | Authors: | Engilberge, S, Ramberg, K, Crowley, P.B. | | Deposit date: | 2020-05-26 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.278 Å) | | Cite: | Facile Fabrication of Protein-Macrocycle Frameworks.
J.Am.Chem.Soc., 143, 2021
|
|
6Z5M
 
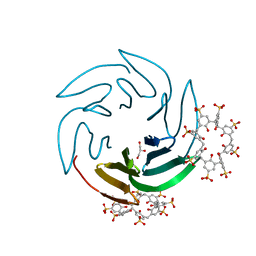 | | The RSL - sulfonato-calix[8]arene complex, I23 form, Gly-HCl pH 2.2 | | Descriptor: | Fucose-binding lectin protein, GLYCEROL, beta-D-fructopyranose, ... | | Authors: | Ramberg, K, Engilberge, S, Crowley, P.B. | | Deposit date: | 2020-05-26 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Facile Fabrication of Protein-Macrocycle Frameworks.
J.Am.Chem.Soc., 143, 2021
|
|
6Z5W
 
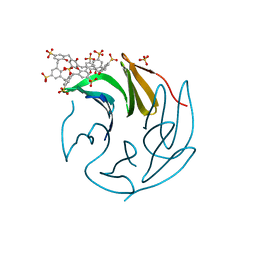 | | The RSL - sulfonato-calix[8]arene complex, P213 form, MES pH 6.8 | | Descriptor: | Fucose-binding lectin protein, SULFATE ION, beta-D-fructopyranose, ... | | Authors: | Engilberge, S, Ramberg, K, Crowley, P.B. | | Deposit date: | 2020-05-27 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.187 Å) | | Cite: | Facile Fabrication of Protein-Macrocycle Frameworks.
J.Am.Chem.Soc., 143, 2021
|
|
6TWN
 
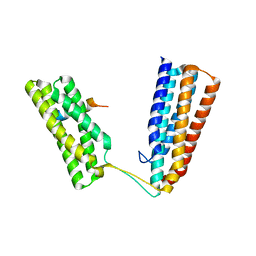 | | Crystal structure of Talin1 R7R8 in complex with CDK1 (206-223) | | Descriptor: | CHLORIDE ION, Cyclin-dependent kinase 1, GLYCEROL, ... | | Authors: | Zacharchenko, T, Muench, S.P, Goult, B.T. | | Deposit date: | 2020-01-13 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Talin mechanosensitivity is modulated by a direct interaction with cyclin-dependent kinase-1.
J.Biol.Chem., 297, 2021
|
|
6Z5Z
 
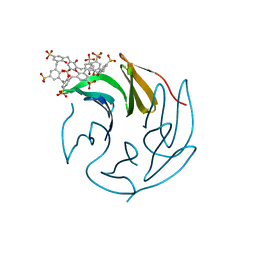 | | The RSL-R6 - sulfonato-calix[8]arene complex, P213 form, TRIS-HCl pH 8.5 | | Descriptor: | Fucose-binding lectin protein, beta-D-fructopyranose, sulfonato-calix[8]arene | | Authors: | Ramberg, K, Engilberge, S, Crowley, P.B. | | Deposit date: | 2020-05-27 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.124 Å) | | Cite: | Facile Fabrication of Protein-Macrocycle Frameworks.
J.Am.Chem.Soc., 143, 2021
|
|
6Z60
 
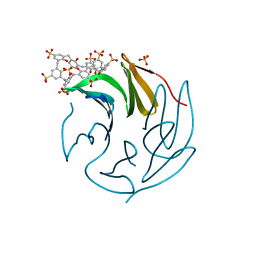 | | The RSL - sulfonato-calix[8]arene complex, P213 form, CAPS pH 9.5 | | Descriptor: | Fucose-binding lectin protein, SULFATE ION, beta-D-fructopyranose, ... | | Authors: | Ramberg, K, Engilberge, S, Crowley, P.B. | | Deposit date: | 2020-05-27 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.166 Å) | | Cite: | Facile Fabrication of Protein-Macrocycle Frameworks.
J.Am.Chem.Soc., 143, 2021
|
|
6Z62
 
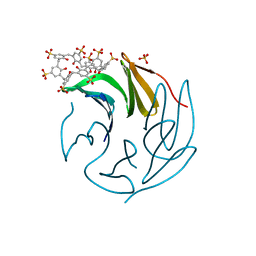 | | The RSL - sulfonato-calix[8]arene complex, P213 form, TRIS-HCl pH 8.5 | | Descriptor: | Fucose-binding lectin protein, SULFATE ION, beta-D-fructopyranose, ... | | Authors: | Ramberg, K, Engilberge, S, Crowley, P.B. | | Deposit date: | 2020-05-27 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.156 Å) | | Cite: | Facile Fabrication of Protein-Macrocycle Frameworks.
J.Am.Chem.Soc., 143, 2021
|
|
