1KH7
 
 | | E. COLI ALKALINE PHOSPHATASE MUTANT (D153GD330N) | | Descriptor: | MAGNESIUM ION, SULFATE ION, ZINC ION, ... | | Authors: | Le Du, M.H, Lamoure, C, Muller, B.H, Bulgakov, O.V, Lajeunesse, E. | | Deposit date: | 2001-11-29 | | Release date: | 2002-03-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Artificial evolution of an enzyme active site: structural studies of three highly active mutants of Escherichia coli alkaline phosphatase.
J.Mol.Biol., 316, 2002
|
|
1KHL
 
 | | E. COLI ALKALINE PHOSPHATASE MUTANT (D153HD330N) COMPLEX WITH PHOSPHATE | | Descriptor: | Alkaline Phosphatase, PHOSPHATE ION, ZINC ION | | Authors: | Le Du, M.H, Lamoure, C, Muller, B.H, Bulgakov, O.V, Lajeunesse, E, Menez, A, Boulain, J.C. | | Deposit date: | 2001-11-30 | | Release date: | 2002-03-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Artificial evolution of an enzyme active site: structural studies of three highly active mutants of Escherichia coli alkaline phosphatase.
J.Mol.Biol., 316, 2002
|
|
5WLY
 
 | | E. coli LpxH- 8 mutations | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Bohl, T.E, Aihara, H, Shi, K, Lee, J.K. | | Deposit date: | 2017-07-28 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The substrate-binding cap of the UDP-diacylglucosamine pyrophosphatase LpxH is highly flexible, enabling facile substrate binding and product release.
J. Biol. Chem., 293, 2018
|
|
4YLN
 
 | | E. coli Transcription Initiation Complex - 17-bp spacer and 4-nt RNA | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zuo, Y, Steitz, T.A. | | Deposit date: | 2015-03-05 | | Release date: | 2015-04-22 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (5.5 Å) | | Cite: | Crystal Structures of the E. coli Transcription Initiation Complexes with a Complete Bubble.
Mol.Cell, 58, 2015
|
|
1KHN
 
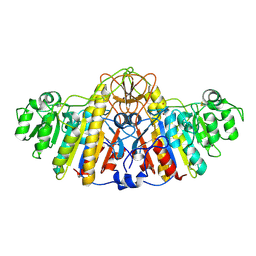 | | E. COLI ALKALINE PHOSPHATASE MUTANT (D153HD330N) ZINC FORM | | Descriptor: | Alkaline phosphatase, ZINC ION | | Authors: | Le Du, M.H, Lamoure, C, Muller, B.H, Bulgakov, O.V, Lajeunesse, E, Menez, A, Boulain, J.C. | | Deposit date: | 2001-11-30 | | Release date: | 2002-03-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Artificial evolution of an enzyme active site: structural studies of three highly active mutants of Escherichia coli alkaline phosphatase.
J.Mol.Biol., 316, 2002
|
|
1KH9
 
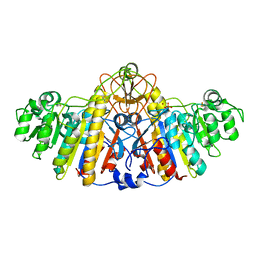 | | E. COLI ALKALINE PHOSPHATASE MUTANT (D153GD330N) COMPLEX WITH PHOSPHATE | | Descriptor: | Alkaline phosphatase, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Le Du, M.H, Lamoure, C, Muller, B.H, Bulgakov, O.V, Lajeunesse, E. | | Deposit date: | 2001-11-29 | | Release date: | 2002-03-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Artificial evolution of an enzyme active site: structural studies of three highly active mutants of Escherichia coli alkaline phosphatase.
J.Mol.Biol., 316, 2002
|
|
1KHK
 
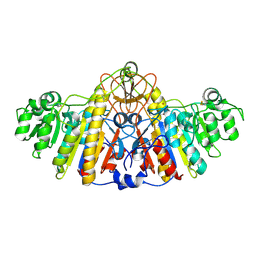 | | E. COLI ALKALINE PHOSPHATASE MUTANT (D153HD330N) | | Descriptor: | Alkaline Phosphatase, MAGNESIUM ION, ZINC ION | | Authors: | Le Du, M.H, Lamoure, C, Muller, B.H, Bulgakov, O.V, Lajeunesse, E, Menez, A, Boulain, J.C. | | Deposit date: | 2001-11-30 | | Release date: | 2002-03-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Artificial evolution of an enzyme active site: structural studies of three highly active mutants of Escherichia coli alkaline phosphatase.
J.Mol.Biol., 316, 2002
|
|
1KH4
 
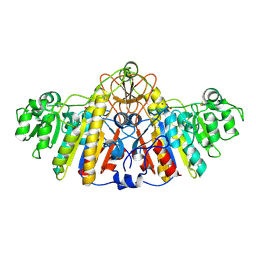 | | E. COLI ALKALINE PHOSPHATASE MUTANT (D330N) IN COMPLEX WITH PHOSPHATE | | Descriptor: | ALKALINE PHOSPHATASE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Le Du, M.H, Lamoure, C, Muller, B.H, Bulgakov, O.V, Lajeunesse, E. | | Deposit date: | 2001-11-29 | | Release date: | 2002-03-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Artificial evolution of an enzyme active site: structural studies of three highly active mutants of Escherichia coli alkaline phosphatase.
J.Mol.Biol., 316, 2002
|
|
7Z15
 
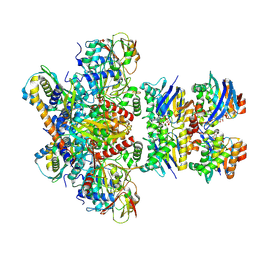 | | E. coli C-P lyase bound to a PhnK/PhnL dual ABC dimer and ADP + Pi | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Alpha-D-ribose 1-methylphosphonate 5-phosphate C-P lyase, ... | | Authors: | Amstrup, S.K, Sofos, N, Karlsen, J.L, Skjerning, R.B, Boesen, T, Enghild, J.J, Hove-Jensen, B, Brodersen, D.E. | | Deposit date: | 2022-02-24 | | Release date: | 2022-06-22 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (1.93 Å) | | Cite: | Structural remodelling of the carbon-phosphorus lyase machinery by a dual ABC ATPase.
Nat Commun, 14, 2023
|
|
7Z16
 
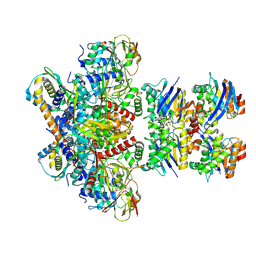 | | E. coli C-P lyase bound to PhnK/PhnL dual ABC dimer with AMPPNP and PhnK E171Q mutation | | Descriptor: | Alpha-D-ribose 1-methylphosphonate 5-phosphate C-P lyase, Alpha-D-ribose 1-methylphosphonate 5-triphosphate synthase subunit PhnH, Alpha-D-ribose 1-methylphosphonate 5-triphosphate synthase subunit PhnI, ... | | Authors: | Amstrup, S.K, Sofus, N, Karlsen, J.L, Skjerning, R.B, Boesen, T, Enghild, J.J, Hove-Jensen, B, Brodersen, D.E. | | Deposit date: | 2022-02-24 | | Release date: | 2022-06-22 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.09 Å) | | Cite: | Structural remodelling of the carbon-phosphorus lyase machinery by a dual ABC ATPase.
Nat Commun, 14, 2023
|
|
1PE6
 
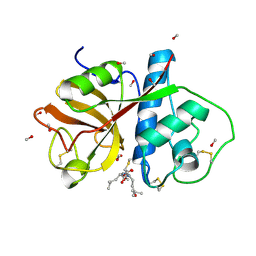 | | REFINED X-RAY STRUCTURE OF PAPAIN(DOT)E-64-C COMPLEX AT 2.1-ANGSTROMS RESOLUTION | | Descriptor: | METHANOL, N-[1-HYDROXYCARBOXYETHYL-CARBONYL]LEUCYLAMINO-2-METHYL-BUTANE, PAPAIN | | Authors: | Yamamoto, D, Matsumoto, K, Ohishi, H, Ishida, T, Inoue, M, Kitamura, K, Mizuno, H. | | Deposit date: | 1991-05-14 | | Release date: | 1993-04-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Refined x-ray structure of papain.E-64-c complex at 2.1-A resolution.
J.Biol.Chem., 266, 1991
|
|
1KH5
 
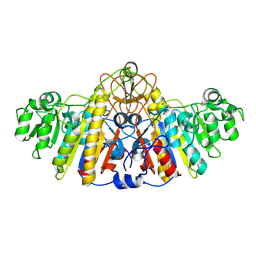 | | E. COLI ALKALINE PHOSPHATASE MUTANT (D330N) MIMIC OF THE TRANSITION STATES WITH ALUMINIUM FLUORIDE | | Descriptor: | ALKALINE PHOSPHATASE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | Authors: | Le Du, M.H, Lamoure, C, Muller, B.H, Bulgakov, O.V, Lajeunesse, E. | | Deposit date: | 2001-11-29 | | Release date: | 2002-03-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Artificial evolution of an enzyme active site: structural studies of three highly active mutants of Escherichia coli alkaline phosphatase.
J.Mol.Biol., 316, 2002
|
|
1KHJ
 
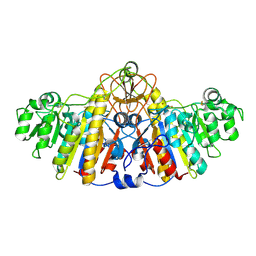 | | E. COLI ALKALINE PHOSPHATASE MUTANT (D153HD330N) MIMIC OF THE TRANSITION STATES WITH ALUMINIUM FLUORIDE | | Descriptor: | ALUMINUM FLUORIDE, Alkaline phosphatase, ZINC ION | | Authors: | Le Du, M.H, Lamoure, C, Muller, B.H, Bulgakov, O.V, Lajeunesse, E, Menez, A, Boulain, J.C. | | Deposit date: | 2001-11-30 | | Release date: | 2002-03-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Artificial evolution of an enzyme active site: structural studies of three highly active mutants of Escherichia coli alkaline phosphatase.
J.Mol.Biol., 316, 2002
|
|
3MUY
 
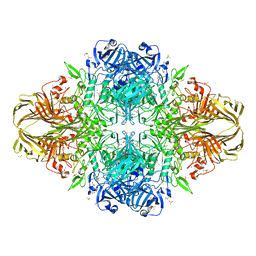 | | E. coli (lacZ) beta-galactosidase (R599A) | | Descriptor: | Beta-D-galactosidase, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Dugdale, M.L, Vance, M, Driedger, M.L, Nibber, A, Tran, A, Huber, R.E. | | Deposit date: | 2010-05-03 | | Release date: | 2011-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Importance of Arg-599 of beta-galactosidase (Escherichia coli) as an anchor for the open conformations of Phe-601 and the active-site loop
Biochem.Cell Biol., 88, 2010
|
|
6P9P
 
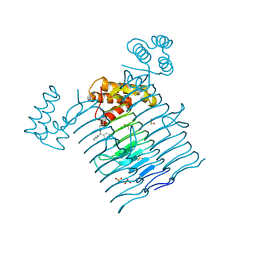 | | E.coli LpxA in complex with Compound 1 | | Descriptor: | 3-[2-(4-methoxyphenyl)-2-oxoethyl]-5,5-diphenylimidazolidine-2,4-dione, Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, DIMETHYL SULFOXIDE, ... | | Authors: | Ma, X, Shia, S, Ornelas, E. | | Deposit date: | 2019-06-10 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Two Distinct Mechanisms of Inhibition of LpxA Acyltransferase Essential for Lipopolysaccharide Biosynthesis.
J.Am.Chem.Soc., 142, 2020
|
|
6P9R
 
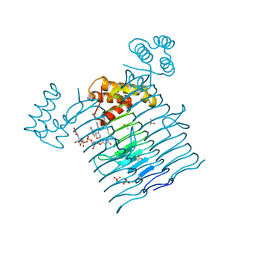 | | E.coli LpxA in complex with UDP-3-O-(R-3-hydroxymyristoyl)-GlcNAc and Compound 6 | | Descriptor: | Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, DIMETHYL SULFOXIDE, PHOSPHATE ION, ... | | Authors: | Ma, X, Shia, S, Ornelas, E. | | Deposit date: | 2019-06-10 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Two Distinct Mechanisms of Inhibition of LpxA Acyltransferase Essential for Lipopolysaccharide Biosynthesis.
J.Am.Chem.Soc., 142, 2020
|
|
6P9T
 
 | | E.coli LpxA in complex with UDP-3-O-(R-3-hydroxymyristoyl)-GlcNAc and Compound 8 | | Descriptor: | Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, DIMETHYL SULFOXIDE, PHOSPHATE ION, ... | | Authors: | Ma, X, Shia, S, Ornelas, E. | | Deposit date: | 2019-06-10 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Two Distinct Mechanisms of Inhibition of LpxA Acyltransferase Essential for Lipopolysaccharide Biosynthesis.
J.Am.Chem.Soc., 142, 2020
|
|
4XKU
 
 | | E coli BFR variant Y114F | | Descriptor: | Bacterioferritin, SULFATE ION | | Authors: | Hemmings, A.M, Le Brun, N.E, Bradley, J.M. | | Deposit date: | 2015-01-12 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Three Aromatic Residues are Required for Electron Transfer during Iron Mineralization in Bacterioferritin.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
7U48
 
 | | Clavulanic acid-CTX-M-15 | | Descriptor: | (E)-5-hydroxy-3-oxo-N-(3-oxopropylidene)-L-norvaline, Beta-lactamase, GLYCEROL, ... | | Authors: | Ahmadvand, P, Kang, C.H. | | Deposit date: | 2022-02-28 | | Release date: | 2022-05-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Characterization of Interactions between CTX-M-15 and Clavulanic Acid, Desfuroylceftiofur, Ceftiofur, Ampicillin, and Nitrocefin.
Int J Mol Sci, 23, 2022
|
|
5FGZ
 
 | | E. coli PBP1b in complex with FPI-1465 | | Descriptor: | MOENOMYCIN, Penicillin-binding protein 1B, [[(3~{R},6~{S})-1-methanoyl-6-[[(3~{S})-pyrrolidin-3-yl]oxycarbamoyl]piperidin-3-yl]amino] hydrogen sulfate | | Authors: | King, D.T, Strynadka, N.C.J. | | Deposit date: | 2015-12-21 | | Release date: | 2016-01-20 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural and Kinetic Characterization of Diazabicyclooctanes as Dual Inhibitors of Both Serine-beta-Lactamases and Penicillin-Binding Proteins.
Acs Chem.Biol., 11, 2016
|
|
3MV1
 
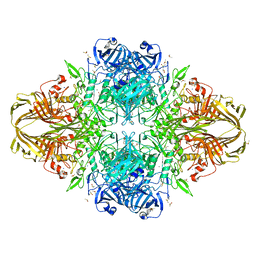 | | E.Coli (lacZ) beta-galactosidase (R599A) in complex with Guanidinium | | Descriptor: | Beta-galactosidase, DIMETHYL SULFOXIDE, GUANIDINE, ... | | Authors: | Dugdale, M.L, Vance, M, Driedger, M.L, Nibber, A, Tran, A, Huber, R.E. | | Deposit date: | 2010-05-03 | | Release date: | 2011-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Importance of Arg-599 of b-galactosidase (Escherichia coli) as an anchor for the open conformations of Phe-601 and the active-site loop
Biochem.Cell Biol., 88, 2010
|
|
2WDQ
 
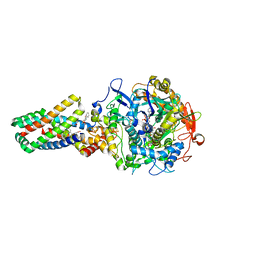 | | E. coli succinate:quinone oxidoreductase (SQR) with carboxin bound | | Descriptor: | 2-METHYL-N-PHENYL-5,6-DIHYDRO-1,4-OXATHIINE-3-CARBOXAMIDE, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Ruprecht, J, Yankovskaya, V, Maklashina, E, Iwata, S, Cecchini, G. | | Deposit date: | 2009-03-25 | | Release date: | 2009-08-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of Escherichia coli succinate:quinone oxidoreductase with an occupied and empty quinone-binding site.
J. Biol. Chem., 284, 2009
|
|
7Z18
 
 | | E. coli C-P lyase bound to a PhnK ABC dimer and ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Alpha-D-ribose 1-methylphosphonate 5-phosphate C-P lyase, Alpha-D-ribose 1-methylphosphonate 5-triphosphate synthase subunit PhnG, ... | | Authors: | Amstrup, S.K, Sofos, N, Karlsen, J.L, Skjerning, R.B, Boesen, T, Enghild, J.J, Hove-Jensen, B, Brodersen, D.E. | | Deposit date: | 2022-02-24 | | Release date: | 2022-05-25 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (1.98 Å) | | Cite: | Structural remodelling of the carbon-phosphorus lyase machinery by a dual ABC ATPase.
Nat Commun, 14, 2023
|
|
7Z19
 
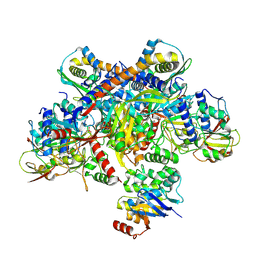 | | E. coli C-P lyase bound to a single PhnK ABC domain | | Descriptor: | Alpha-D-ribose 1-methylphosphonate 5-phosphate C-P lyase, Alpha-D-ribose 1-methylphosphonate 5-triphosphate synthase subunit PhnG, Alpha-D-ribose 1-methylphosphonate 5-triphosphate synthase subunit PhnH, ... | | Authors: | Amstrup, S.K, Sofos, N, Karlsen, J.L, Skjerning, R.B, Boesen, T, Enghild, J.J, Hove-Jensen, B, Brodersen, D.E. | | Deposit date: | 2022-02-24 | | Release date: | 2022-05-25 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Structural remodelling of the carbon-phosphorus lyase machinery by a dual ABC ATPase.
Nat Commun, 14, 2023
|
|
7Z17
 
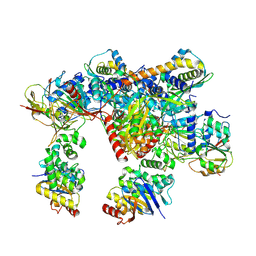 | | E. coli C-P lyase bound to a PhnK ABC dimer in an open conformation | | Descriptor: | Alpha-D-ribose 1-methylphosphonate 5-phosphate C-P lyase, Alpha-D-ribose 1-methylphosphonate 5-triphosphate synthase subunit PhnG, Alpha-D-ribose 1-methylphosphonate 5-triphosphate synthase subunit PhnH, ... | | Authors: | Amstrup, S.K, Sofos, N, Karlsen, J.L, Skjerning, R.B, Boesen, T, Enghild, J.J, Hove-Jensen, B, Brodersen, D.E. | | Deposit date: | 2022-02-24 | | Release date: | 2022-05-25 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Structural remodelling of the carbon-phosphorus lyase machinery by a dual ABC ATPase.
Nat Commun, 14, 2023
|
|
