3UAU
 
 | | Crystal structure of the lipoprotein JlpA | | Descriptor: | Surface-exposed lipoprotein | | Authors: | Kawai, F, Yeo, H.J. | | Deposit date: | 2011-10-22 | | Release date: | 2012-07-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of JlpA, a surface-exposed lipoprotein adhesin of Campylobacter jejuni.
J.Struct.Biol., 177, 2012
|
|
1D1D
 
 | |
3UGS
 
 | | Crystal structure of a probable undecaprenyl diphosphate synthase (uppS) from Campylobacter jejuni | | Descriptor: | (2Z,6Z)-3,7,11-trimethyldodeca-2,6,10-trien-1-yl dihydrogen phosphate, Undecaprenyl pyrophosphate synthase | | Authors: | Nocek, B, Gu, M, Grimshaw, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-11-02 | | Release date: | 2011-11-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.457 Å) | | Cite: | Crystal structure of a probable undecaprenyl diphosphate synthase (uppS) from Campylobacter jejuni
TO BE PUBLISHED
|
|
4R53
 
 | |
3TZL
 
 | | Crystal Structure of Tryptophanyl-tRNA Synthetase from Campylobacter jejuni complexed with ADP and Tryptophane | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, PHOSPHATE ION, SODIUM ION, ... | | Authors: | Kim, Y, Zhou, M, Grimshaw, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-09-27 | | Release date: | 2011-10-05 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.154 Å) | | Cite: | Crystal Structure of Tryptophanyl-tRNA Synthetase from Campylobacter jejuni complexed with ADP and Tryptophane
To be Published
|
|
3UDO
 
 | | Crystal structure of putative isopropylamlate dehydrogenase from Campylobacter jejuni | | Descriptor: | 1,2-ETHANEDIOL, 3-isopropylmalate dehydrogenase, SULFATE ION | | Authors: | Tkaczuk, K.L, Chruszcz, M, Blus, B.J, Onopriyenko, O, Grimshaw, S, Savchenko, A, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-10-28 | | Release date: | 2011-11-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of putative isopropylamlate dehydrogenase from Campylobacter jejuni
To be Published
|
|
4R7J
 
 | | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase with the Internal Deletion Containing CBS Domain from Campylobacter jejuni | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Gu, M, Hedstrom, L, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-08-27 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1172 Å) | | Cite: | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase with the Internal Deletion Containing CBS Domain from Campylobacter jejuni
To be Published, 2014
|
|
1G6E
 
 | | ANTIFUNGAL PROTEIN FROM STREPTOMYCES TENDAE TU901, 30-CONFORMERS ENSEMBLE | | Descriptor: | ANTIFUNGAL PROTEIN | | Authors: | Campos-Olivas, R, Bormann, C, Hoerr, I, Jung, G, Gronenborn, A.M. | | Deposit date: | 2000-11-04 | | Release date: | 2001-03-28 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Solution structure, backbone dynamics and chitin binding of the anti-fungal protein from Streptomyces tendae TU901.
J.Mol.Biol., 308, 2001
|
|
3KWO
 
 | | Crystal Structure of Putative Bacterioferritin from Campylobacter jejuni | | Descriptor: | 1,4-BUTANEDIOL, ACETIC ACID, GLYCEROL, ... | | Authors: | Kim, Y, Gu, M, Papazisi, L, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-12-01 | | Release date: | 2010-01-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.985 Å) | | Cite: | Crystal Structure of Putative Bacterioferritin from Campylobacter jejuni
To be Published
|
|
2AKO
 
 | |
4ZO4
 
 | | Dephospho-CoA kinase from Campylobacter jejuni. | | Descriptor: | BETA-MERCAPTOETHANOL, Dephospho-CoA kinase | | Authors: | Osipiuk, J, Zhou, M, Stam, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-05-06 | | Release date: | 2015-05-13 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Dephospho-CoA kinase from Campylobacter jejuni.
to be published
|
|
5UQF
 
 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Campylobacter jejuni in the complex with IMP and the inhibitor P225 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Kim, Y, Maltseva, N, Makowska-Grzyska, M, Gu, M, Gollapalli, D, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-08 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from
Campylobacter jejuni in the complex with IMP and the inhibitor P225
To Be Published
|
|
5URQ
 
 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Campylobacter jejuni in the complex with inhibitor p176 | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase, N-{2-chloro-5-[({2-[3-(prop-1-en-2-yl)phenyl]propan-2-yl}carbamoyl)amino]phenyl}-alpha-D-ribofuranosylamine, ... | | Authors: | Kim, Y, Maltseva, N, Makowska-Grzyska, M, Gu, M, Gollapalli, D, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-12 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Campylobacter jejuni in the complex with inhibitor p176
To Be Published
|
|
1EWA
 
 | | Dehaloperoxidase and 4-iodophenol | | Descriptor: | 4-IODOPHENOL, DEHALOPEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | LaCount, M.W, Zhang, E, Chen, Y.P, Han, K, Whitton, M.M, Lincoln, D.E, Woodin, S.A, Lebioda, L. | | Deposit date: | 2000-04-24 | | Release date: | 2000-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure and amino acid sequence of dehaloperoxidase from Amphitrite ornata indicate common ancestry with globins
J.Biol.Chem., 275, 2000
|
|
3L84
 
 | | High resolution crystal structure of transketolase from Campylobacter jejuni subsp. jejuni NCTC 11168 | | Descriptor: | ACETATE ION, GLYCEROL, Transketolase | | Authors: | Nocek, B, Makowska-Grzyska, M, Maltseva, N, Grimshaw, S, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-12-29 | | Release date: | 2010-02-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | High resolution crystal structure of transketolase from Campylobacter jejuni subsp. jejuni NCTC 11168
To be Published
|
|
4FH7
 
 | | Structure of DHP A in complex with 2,4,6-tribromophenol in 20% methanol | | Descriptor: | 2,4,6-TRIBROMOPHENOL, Dehaloperoxidase A, METHANOL, ... | | Authors: | de Serrano, V.S, Franzen, S. | | Deposit date: | 2012-06-05 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural and Kinetic Study of an Internal Substrate Binding Site in Dehaloperoxidase-Hemoglobin A from Amphitrite ornata.
Biochemistry, 52, 2013
|
|
3MOU
 
 | |
2FGS
 
 | | Crystal structure of Campylobacter jejuni YCEI protein, structural genomics | | Descriptor: | Putative periplasmic protein, SULFATE ION | | Authors: | Patskovsky, Y, Ramagopal, U, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-12-22 | | Release date: | 2006-01-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of Campylobacter Jejuni YceI Periplasmic Protein
To be Published
|
|
3M5V
 
 | | Crystal Structure of Dihydrodipicolinate Synthase from Campylobacter jejuni | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Dihydrodipicolinate synthase, ... | | Authors: | Kim, Y, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-03-13 | | Release date: | 2010-04-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Dihydrodipicolinate Synthase from Campylobacter jejuni
To be Published
|
|
3M5U
 
 | | Crystal Structure of Phosphoserine Aminotransferase from Campylobacter jejuni | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, Phosphoserine aminotransferase | | Authors: | Kim, Y, Gu, M, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-03-13 | | Release date: | 2010-04-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.152 Å) | | Cite: | Crystal Structure of Phosphoserine Aminotransferase from Campylobacter jejuni
To be Published
|
|
5UQG
 
 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Campylobacter jejuni in the complex with inhibitor p200 | | Descriptor: | 1,2-ETHANEDIOL, 3-(2-{[(4-chlorophenyl)carbamoyl]amino}propan-2-yl)-N-hydroxybenzene-1-carboximidamide, INOSINIC ACID, ... | | Authors: | Kim, Y, Maltseva, N, Makowska-Grzyska, M, Gu, M, Gollapalli, D, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-08 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Campylobacter jejuni in the complex with inhibitor p200
To Be Published
|
|
2ADX
 
 | | FIFTH EGF-LIKE DOMAIN OF THROMBOMODULIN (TMEGF5), NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | THROMBOMODULIN | | Authors: | Sampoli-Benitez, B.A, Hunter, M.J, Meininger, D.P, Komives, E.A. | | Deposit date: | 1997-02-18 | | Release date: | 1997-12-24 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Structure of the fifth EGF-like domain of thrombomodulin: An EGF-like domain with a novel disulfide-bonding pattern.
J.Mol.Biol., 273, 1997
|
|
3THA
 
 | | Tryptophan synthase subunit alpha from Campylobacter jejuni. | | Descriptor: | Tryptophan synthase alpha chain | | Authors: | Osipiuk, J, Gu, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-08-18 | | Release date: | 2011-08-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Tryptophan synthase subunit alpha from Campylobacter jejuni.
To be Published
|
|
5CHQ
 
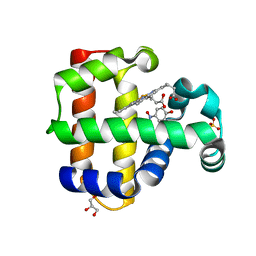 | | Dehaloperoxidase B in complex with substrate p-nitrophenol | | Descriptor: | Dehaloperoxidase B, GLYCEROL, P-NITROPHENOL, ... | | Authors: | Carey, L, Ghiladi, R. | | Deposit date: | 2015-07-10 | | Release date: | 2016-04-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Nonmicrobial Nitrophenol Degradation via Peroxygenase Activity of Dehaloperoxidase-Hemoglobin from Amphitrite ornata.
Biochemistry, 55, 2016
|
|
5CHR
 
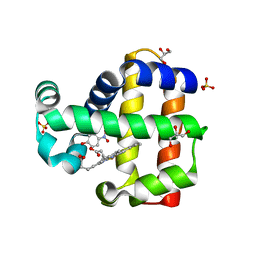 | | Dehaloperoxidase B in complex with substrate p-nitrocatechol | | Descriptor: | 4-NITROCATECHOL, Dehaloperoxidase B, GLYCEROL, ... | | Authors: | Carey, L, Ghiladi, R. | | Deposit date: | 2015-07-10 | | Release date: | 2016-04-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Nonmicrobial Nitrophenol Degradation via Peroxygenase Activity of Dehaloperoxidase-Hemoglobin from Amphitrite ornata.
Biochemistry, 55, 2016
|
|
