1OLG
 
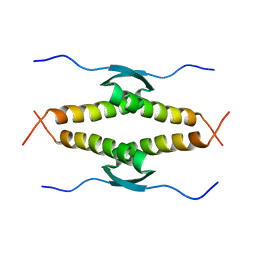 | |
1QVX
 
 | | SOLUTION STRUCTURE OF THE FAT DOMAIN OF FOCAL ADHESION KINASE | | Descriptor: | Focal adhesion kinase 1 | | Authors: | Gao, G, Prutzman, K.C, King, M.L, DeRose, E.F, London, R.E, Schaller, M.D, Campbell, S.L. | | Deposit date: | 2003-08-29 | | Release date: | 2004-03-02 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of the Focal Adhesion Targeting Domain of Focal Adhesion Kinase in Complex with a Paxillin LD Peptide: EVIDENCE FOR A TWO-SITE BINDING MODEL.
J.Biol.Chem., 279, 2004
|
|
1CCM
 
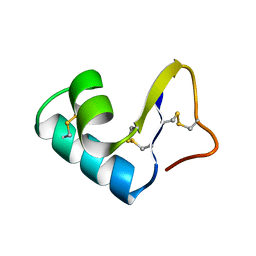 | | DIRECT NOE REFINEMENT OF CRAMBIN FROM 2D NMR DATA USING A SLOW-COOLING ANNEALING PROTOCOL | | Descriptor: | CRAMBIN | | Authors: | Bonvin, A.M.J.J, Rullmann, J.A.C, Lamerichs, R.M.J.N, Boelens, R, Kaptein, R. | | Deposit date: | 1993-04-14 | | Release date: | 1993-10-31 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | "Ensemble" iterative relaxation matrix approach: a new NMR refinement protocol applied to the solution structure of crambin.
Proteins, 15, 1993
|
|
1EQ8
 
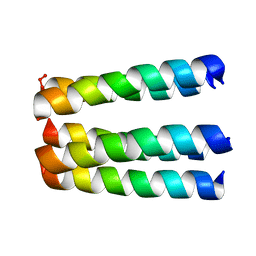 | | THREE-DIMENSIONAL STRUCTURE OF THE PENTAMERIC HELICAL BUNDLE OF THE ACETYLCHOLINE RECEPTOR M2 TRANSMEMBRANE SEGMENT | | Descriptor: | ACETYLCHOLINE RECEPTOR PROTEIN, HYDROXIDE ION | | Authors: | Marassi, F.M, Gesell, J.J, Kim, Y, Valente, A.P, Oblatt-Montal, M, Montal, M, Opella, S.J. | | Deposit date: | 2000-04-03 | | Release date: | 2000-04-26 | | Last modified: | 2022-02-16 | | Method: | SOLID-STATE NMR | | Cite: | Structures of the M2 channel-lining segments from nicotinic acetylcholine and NMDA receptors by NMR spectroscopy.
Nat.Struct.Biol., 6, 1999
|
|
1IDX
 
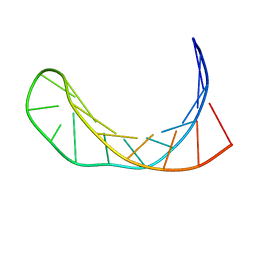 | |
1RYJ
 
 | | Solution NMR Structure of Protein Mth1743 from Methanobacterium thermoautotrophicum. Ontario Centre for Structural Proteomics target MTH1743_1_70; Northeast Structural Genomics Consortium Target TT526. | | Descriptor: | unknown | | Authors: | Yee, A, Chang, X, Pineda-Lucena, A, Wu, B, Semesi, A, Le, B, Ramelot, T, Lee, G.M, Bhattacharyya, S, Gutierrez, P, Denisov, A, Lee, C.H, Cort, J.R, Kozlov, G, Liao, J, Finak, G, Chen, L, Wishart, D, Lee, W, McIntosh, L.P, Gehring, K, Kennedy, M.A, Edwards, A.M, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-12-22 | | Release date: | 2004-02-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | AN NMR APPROACH TO STRUCTURAL PROTEOMICS
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1SX1
 
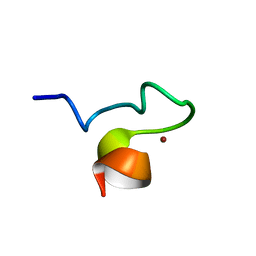 | | Solution NMR Structure and X-ray Absorption Analysis of the C-Terminal Zinc-Binding Domain of the SecA ATPase | | Descriptor: | SecA, ZINC ION | | Authors: | Dempsey, B.R, Wrona, M, Moulin, J.M, Gloor, G.B, Jalilehvand, F, Lajoie, G, Shaw, G.S, Shilton, B.H. | | Deposit date: | 2004-03-30 | | Release date: | 2004-07-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure and X-ray Absorption Analysis of the C-Terminal Zinc-Binding Domain of the SecA ATPase.
Biochemistry, 43, 2004
|
|
5IRT
 
 | |
1EZB
 
 | |
1EZA
 
 | |
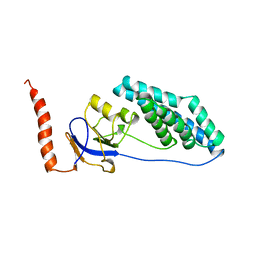
1EZD
 
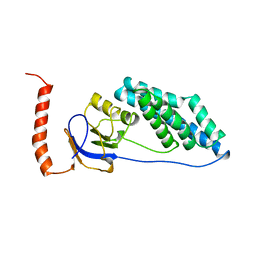 | |
1EZC
 
 | |
1SX0
 
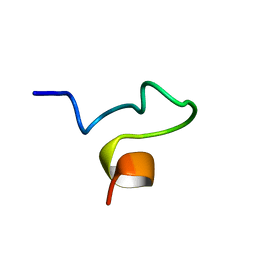 | | Solution NMR Structure and X-Ray Absorption Analysis of the C-Terminal Zinc-Binding Domain of the SecA ATPase | | Descriptor: | SecA | | Authors: | Dempsey, B.R, Wrona, M, Moulin, J.M, Gloor, G.B, Jalilehvand, F, Lajoie, G, Shaw, G.S, Shilton, B.H. | | Deposit date: | 2004-03-30 | | Release date: | 2004-07-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure and X-ray Absorption Analysis of the C-Terminal Zinc-Binding Domain of the SecA ATPase.
Biochemistry, 43, 2004
|
|
1IG6
 
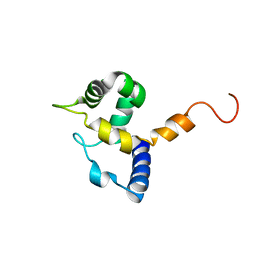 | | HUMAN MRF-2 DOMAIN, NMR, 11 STRUCTURES | | Descriptor: | MODULATOR RECOGNITION FACTOR 2 | | Authors: | Lin, D, Tsui, V, Case, D, Yuan, Y.C, Chen, Y. | | Deposit date: | 2001-04-17 | | Release date: | 2001-04-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | HUMAN MRF-2 DOMAIN, NMR, 11 STRUCTURES
To be Published
|
|
1MR0
 
 | | SOLUTION NMR STRUCTURE OF AGRP(87-120; C105A) | | Descriptor: | AGOUTI RELATED PROTEIN | | Authors: | Jackson, P.J, Mcnulty, J.C, Yang, Y.K, Thompson, D.A, Chai, B, Gantz, I, Barsh, G.S, Millhauser, G.M. | | Deposit date: | 2002-09-17 | | Release date: | 2002-10-02 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Design, pharmacology, and NMR structure of a minimized cystine knot with agouti-related protein activity.
Biochemistry, 41, 2002
|
|
1F22
 
 | | A PROTON-NMR INVESTIGATION OF THE FULLY REDUCED CYTOCHROME C7 FROM DESULFUROMONAS ACETOXIDANS. COMPARISON BETWEEN THE REDUCED AND THE OXIDIZED FORMS. | | Descriptor: | CYTOCHROME C7, HEME C | | Authors: | Assfalg, M, Banci, L, Bertini, I, Bruschi, M, Giudici-Orticoni, M.T. | | Deposit date: | 2000-05-23 | | Release date: | 2000-06-21 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | A proton-NMR investigation of the fully reduced cytochrome c7 from Desulfuromonas acetoxidans. Comparison between the reduced and the oxidized forms.
Eur.J.Biochem., 266, 1999
|
|
1M9W
 
 | |
5ZNF
 
 | |
1OLH
 
 | |
1HM1
 
 | | THE SOLUTION NMR STRUCTURE OF A THERMALLY STABLE FAPY ADDUCT OF AFLATOXIN B1 IN AN OLIGODEOXYNUCLEOTIDE DUPLEX REFINED FROM DISTANCE RESTRAINED MOLECULAR DYNAMICS SIMULATED ANNEALING, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | DNA (5'-D(*CP*TP*AP*TP*(FAG)P*AP*TP*TP*CP*A)-3'), DNA (5'-D(TP*GP*AP*AP*TP*CP*AP*TP*AP*G)-3') | | Authors: | Mao, H, Deng, Z, Wang, F, Harris, T.M, Stone, M.P. | | Deposit date: | 1998-05-11 | | Release date: | 1998-10-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | An intercalated and thermally stable FAPY adduct of aflatoxin B1 in a DNA duplex: structural refinement from 1H NMR.
Biochemistry, 37, 1998
|
|
1G2T
 
 | |
1G2S
 
 | |
6QBJ
 
 | |
1MWN
 
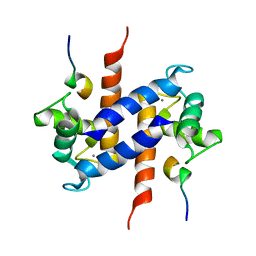 | | Solution NMR structure of S100B bound to the high-affinity target peptide TRTK-12 | | Descriptor: | CALCIUM ION, F-actin capping protein alpha-1 subunit, S-100 protein, ... | | Authors: | Inman, K.G, Yang, R, Rustandi, R.R, Miller, K.E, Baldisseri, D.M, Weber, D.J. | | Deposit date: | 2002-09-30 | | Release date: | 2002-12-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of S100B bound to the high-affinity target peptide TRTK-12
J.Mol.Biol., 324, 2002
|
|
1COP
 
 | |
