6P2D
 
 | | Structure of mouse ketohexokinase-C in complex with fructose and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Ketohexokinase, NITRATE ION, ... | | Authors: | Gasper, W.C, Allen, K.N, Tolan, D.R. | | Deposit date: | 2019-05-21 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Michaelis-like complex of mouse ketohexokinase isoform C
ACTA CRYSTALLOGR.,SECT.D, 2024
|
|
4CJO
 
 | | Spectroscopically-validated structure of ferrous cytochrome c prime from Alcaligenes xylosoxidans, reduced at 180K using X-rays | | Descriptor: | CYTOCHROME C', HEME C | | Authors: | Kekilli, D, Dworkowski, F, Antonyuk, S, Hough, M.A. | | Deposit date: | 2013-12-21 | | Release date: | 2014-05-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Fingerprinting Redox and Ligand States in Haemprotein Crystal Structures Using Resonance Raman Spectroscopy.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3S0E
 
 | | Apis mellifera OBP14 in complex with the odorant eugenol (2-methoxy-4(2-propenyl)-phenol) | | Descriptor: | 2-methoxy-4-(prop-2-en-1-yl)phenol, OBP14 | | Authors: | Spinelli, S, Lagarde, A, Iovinella, I, Tegoni, M, Pelosi, P, Cambillau, C. | | Deposit date: | 2011-05-13 | | Release date: | 2011-11-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of Apis mellifera OBP14, a C-minus odorant-binding protein, and its complexes with odorant molecules.
Insect Biochem.Mol.Biol., 42, 2012
|
|
3S0F
 
 | | Apis mellifera OBP14 native apo, crystal form 2 | | Descriptor: | OBP14 | | Authors: | Spinelli, S, Lagarde, A, Iovinella, I, Tegoni, M, Pelosi, P, Cambillau, C. | | Deposit date: | 2011-05-13 | | Release date: | 2011-11-30 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structure of Apis mellifera OBP14, a C-minus odorant-binding protein, and its complexes with odorant molecules.
Insect Biochem.Mol.Biol., 42, 2012
|
|
3S0G
 
 | | Apis mellifera OBP 14 double mutant Gln44Cys, His97Cys | | Descriptor: | OBP14 | | Authors: | Spinelli, S, Lagarde, A, Iovinella, I, Tegoni, M, Pelosi, P, Cambillau, C. | | Deposit date: | 2011-05-13 | | Release date: | 2011-11-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of Apis mellifera OBP14, a C-minus odorant-binding protein, and its complexes with odorant molecules.
Insect Biochem.Mol.Biol., 42, 2012
|
|
6PFM
 
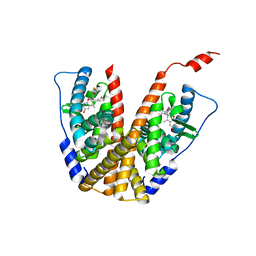 | | Crystal structure of GDC-0927 bound to estrogen receptor alpha | | Descriptor: | (2S)-2-(4-{2-[3-(fluoromethyl)azetidin-1-yl]ethoxy}phenyl)-3-(3-hydroxyphenyl)-4-methyl-2H-1-benzopyran-6-ol, Estrogen receptor | | Authors: | Kiefer, J.R, Vinogradova, M, Liang, J, Zhang, B, Wang, X, Zbieg, J.R, Labadie, S.S, Li, J, Ray, N.C, Ortwine, D. | | Deposit date: | 2019-06-21 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Discovery of a C-8 hydroxychromene as a potent degrader of estrogen receptor alpha with improved rat oral exposure over GDC-0927.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
2QSR
 
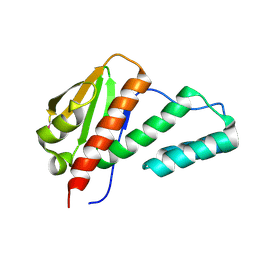 | | Crystal structure of C-terminal domain of transcription-repair coupling factor | | Descriptor: | Transcription-repair coupling factor | | Authors: | Ramagopal, U.A, Toro, R, Gilmore, M, Bain, K, Iizuka, M, Wasserman, S, Rodgers, L, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-07-31 | | Release date: | 2007-09-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of C-terminal domain of transcription-repair coupling factor.
To be Published
|
|
2QSA
 
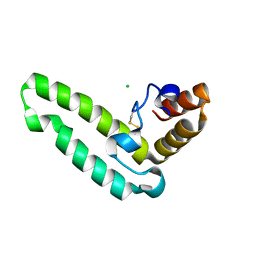 | | Crystal structure of J-domain of DnaJ homolog dnj-2 precursor from C.elegans. | | Descriptor: | CHLORIDE ION, DnaJ homolog dnj-2 | | Authors: | Osipiuk, J, Mulligan, R, Gu, M, Voisine, C, Morimoto, R.I, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-30 | | Release date: | 2007-08-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | X-ray crystal structure of J-domain of DnaJ homolog dnj-2 precursor from C.elegans.
To be Published
|
|
4CG4
 
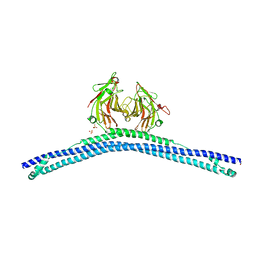 | | Crystal structure of the CHS-B30.2 domains of TRIM20 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-ETHANETHIOL, PYRIN, ... | | Authors: | Weinert, C, Morger, D, Djekic, A, Mittl, P.R.E, Gruetter, M.G. | | Deposit date: | 2013-11-20 | | Release date: | 2015-04-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Trim20 C-Terminal Coiled-Coil/B30.2 Fragment: Implications for the Recognition of Higher Order Oligomers
Sci.Rep., 5, 2015
|
|
4CJG
 
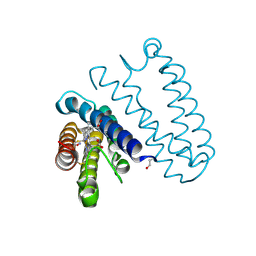 | | Spectroscopically validated structure of the 5 coordinate proximal NO adduct of cytochrome c prime from Alcaligenes xylosoxidans | | Descriptor: | CYTOCHROME C', HEME C, NITRIC OXIDE | | Authors: | Kekilli, D, Dworkowski, F, Fuchs, M, Antonyuk, S, Hough, M.A. | | Deposit date: | 2013-12-20 | | Release date: | 2014-05-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Fingerprinting Redox and Ligand States in Haemprotein Crystal Structures Using Resonance Raman Spectroscopy.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5D9G
 
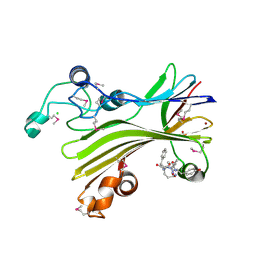 | | Crystal structure of TIPRL, TOR signaling pathway regulator-like, in complex with peptide | | Descriptor: | CHLORIDE ION, NICKEL (II) ION, PHOSPHATE ION, ... | | Authors: | Scorsato, V, Sandy, J, Brandao-Neto, J, Pereira, H.M, Smetana, J.H.C, Aparicio, R. | | Deposit date: | 2015-08-18 | | Release date: | 2016-08-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of the human Tip41 orthologue, TIPRL, reveals a novel fold and a binding site for the PP2Ac C-terminus.
Sci Rep, 6, 2016
|
|
9F2H
 
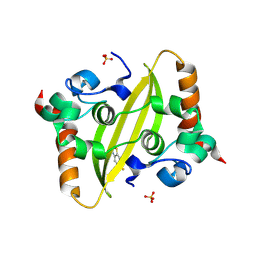 | | Crystal structure of SARS-CoV-2 N-protein C-terminal domain in complex with riluzole | | Descriptor: | 1,2-ETHANEDIOL, 6-(trifluoromethoxy)-1,3-benzothiazol-2-amine, Nucleoprotein, ... | | Authors: | Marquez-Monino, M.A, Gonzalez, B, Perez-Canadillas, J.M. | | Deposit date: | 2024-04-23 | | Release date: | 2024-11-13 | | Last modified: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The ALS drug riluzole binds to the C-terminal domain of SARS-CoV-2 nucleocapsid protein and has antiviral activity.
Structure, 33, 2025
|
|
2G1L
 
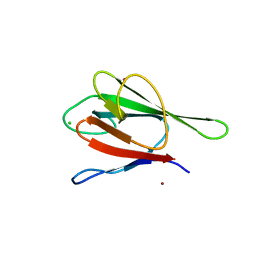 | | Crystal structure of the FHA domain of human kinesin family member C | | Descriptor: | CHLORIDE ION, Kinesin-like protein KIF1C, NICKEL (II) ION, ... | | Authors: | Wang, J, Tempel, W, Shen, Y, Shen, L, Arrowsmith, C, Edwards, A, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-02-14 | | Release date: | 2006-02-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | Crystal structure of the FHA domain of human kinesin family member C
to be published
|
|
9FBK
 
 | |
4CIP
 
 | | Spectroscopically-validated structure of ferrous cytochrome c prime from Alcaligenes xylosoxidans, reduced using ascorbate | | Descriptor: | ASCORBIC ACID, CYTOCHROME C', HEME C, ... | | Authors: | Kekilli, D, Dworkowski, F, Antonyuk, S, Hough, M.A. | | Deposit date: | 2013-12-13 | | Release date: | 2014-05-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Fingerprinting Redox and Ligand States in Haemprotein Crystal Structures Using Resonance Raman Spectroscopy.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
9ESA
 
 | | Aurora-C with SER mutation in complex with INCENP peptide | | Descriptor: | 1,2-ETHANEDIOL, Aurora kinase C, Inner centromere protein | | Authors: | Hillig, R.C. | | Deposit date: | 2024-03-26 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Surface-mutagenesis strategies to enable structural biology crystallization platforms.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
4D4N
 
 | | Nitrosyl complex of the D121A variant of cytochrome c prime from Alcaligenes xylosoxidans | | Descriptor: | CYTOCHROME C', HEME C, NITRIC OXIDE | | Authors: | Gahfoor, D.D, Kekilli, D, Abdullah, G.H, Dworkowski, F.S.N, Hassan, H.G, Wilson, M.T, Hough, M.A, Strange, R.W. | | Deposit date: | 2014-10-30 | | Release date: | 2015-09-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Hydrogen Bonding of the Dissociated Histidine Ligand is not Required for Formation of a Proximal No Adduct in Cytochrome C'.
J.Biol.Inorg.Chem., 20, 2015
|
|
7S0P
 
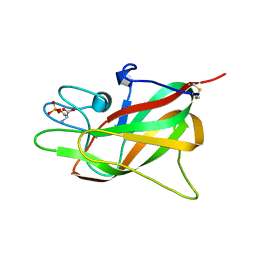 | | Crystal structure of Porcine Factor VIII C2 Domain Bound to Phosphatidylserine | | Descriptor: | Coagulation factor VIII, PHOSPHOSERINE | | Authors: | Peters, S.C, Childers, K.C, Wo, S.W, Brison, C.M, Swanson, C.D, Spiegel, P.C. | | Deposit date: | 2021-08-30 | | Release date: | 2021-10-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Stable binding to phosphatidylserine-containing membranes requires conserved arginine residues in tandem C domains of blood coagulation factor VIII.
Front Mol Biosci, 9, 2022
|
|
9F2I
 
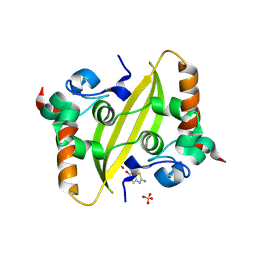 | | Crystal structure of SARS-CoV-2 N-protein C-terminal domain in complex with 2-amino-1,3-benzothiazol-6-ol | | Descriptor: | 1,2-ETHANEDIOL, 2-amino-1,3-benzothiazol-6-ol, Nucleoprotein, ... | | Authors: | Marquez-Monino, M.A, Gonzalez, B, Perez-Canadillas, J.M. | | Deposit date: | 2024-04-23 | | Release date: | 2024-11-13 | | Last modified: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The ALS drug riluzole binds to the C-terminal domain of SARS-CoV-2 nucleocapsid protein and has antiviral activity.
Structure, 33, 2025
|
|
4D4X
 
 | | Nitrosyl complex of the D121I variant of cytochrome c prime from Alcaligenes xylosoxidans | | Descriptor: | CYTOCHROME C', HEME C, NITRIC OXIDE | | Authors: | Gahfoor, D.D, Kekilli, D, Abdullah, G.H, Dworkowski, F.S.N, Hassan, H.G, Wilson, M.T, Hough, M.A, Strange, R.W. | | Deposit date: | 2014-10-31 | | Release date: | 2015-09-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Hydrogen Bonding of the Dissociated Histidine Ligand is not Required for Formation of a Proximal No Adduct in Cytochrome C'.
J.Biol.Inorg.Chem., 20, 2015
|
|
2QKP
 
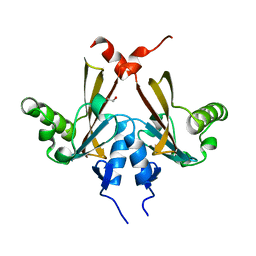 | | Crystal structure of C-terminal domain of SMU_1151c from Streptococcus mutans | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Uncharacterized protein | | Authors: | Ramagopal, U.A, Toro, R, Gilmore, M, Wu, B, Bain, K, Gheyi, T, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-07-11 | | Release date: | 2007-07-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of C-terminal domain of SMU_1151c from Streptococcus mutans.
To be Published
|
|
2QDQ
 
 | | Crystal structure of the talin dimerisation domain | | Descriptor: | Talin-1 | | Authors: | Gingras, A.R, Putz, N.S.M, Bate, N, Barsukov, I.L, Critchley, D.R.C. | | Deposit date: | 2007-06-21 | | Release date: | 2008-01-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of the C-terminal actin-binding domain of talin.
Embo J., 27, 2008
|
|
1WVN
 
 | |
3BRP
 
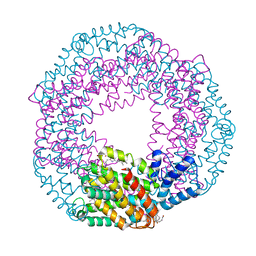 | | Crystal Structure of C-Phycocyanin from Galdieria sulphuraria | | Descriptor: | BILIVERDINE IX ALPHA, C-phycocyanin alpha chain, C-phycocyanin beta chain | | Authors: | Fromme, R. | | Deposit date: | 2007-12-21 | | Release date: | 2009-02-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of C-Phycocyanin from Galdieria sulphuraria at 1.85 A
To be Published
|
|
2QW6
 
 | | Crystal structure of the C-terminal domain of an AAA ATPase from Enterococcus faecium DO | | Descriptor: | AAA ATPase, central region | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Ozyurt, S, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-08-09 | | Release date: | 2007-08-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the C-terminal domain of an AAA ATPase from Enterococcus faecium DO.
To be Published
|
|
