7S7U
 
 | | Crystal structure of iNicSnFR3a Fluorescent Nicotine Sensor with nicotine bound | | Descriptor: | iNicSnFR 3.0 Fluorescent Nicotine Sensor | | Authors: | Fan, C, Shivange, A.V, Looger, L.L, Lester, H.A, Rees, D.C. | | Deposit date: | 2021-09-17 | | Release date: | 2021-10-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Correction: Fluorescence activation mechanism and imaging of drug permeation with new sensors for smoking-cessation ligands.
Elife, 11, 2022
|
|
7S7X
 
 | | Crystal structure of iCytSnFR Cytisine Sensor precursor binding protein with varenicline bound | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, DI(HYDROXYETHYL)ETHER, VARENICLINE, ... | | Authors: | Fan, C, Nichols, N.L, Luebbert, L, Looger, L.L, Lester, H.A, Rees, D.C. | | Deposit date: | 2021-09-17 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure, Function, and Application of Bacterial ABC Transporters
Ph.D.Thesis,California Institute of Technology, 2020
|
|
3U0L
 
 | | Crystal structure of the engineered fluorescent protein mRuby, crystal form 1, pH 4.5 | | Descriptor: | ACETATE ION, mRuby | | Authors: | Akerboom, J, Looger, L.L, Schreiter, E.R. | | Deposit date: | 2011-09-28 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Genetically encoded calcium indicators for multi-color neural activity imaging and combination with optogenetics.
Front Mol Neurosci, 6, 2013
|
|
8SV0
 
 | | The crystal structure of the classical binding interface of Importin alpha 2 and nuclear localisation signal sequence in Psittacine siadenovirus core protein VII | | Descriptor: | Importin subunit alpha-1, SODIUM ION, protein VII | | Authors: | Athukorala, A, Sarker, S, Forwood, J.K, Donnelly, C.M. | | Deposit date: | 2023-05-14 | | Release date: | 2023-05-31 | | Last modified: | 2024-12-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional characterization of siadenovirus core protein VII nuclear localization demonstrates the existence of multiple nuclear transport pathways.
J.Gen.Virol., 105, 2024
|
|
8TGP
 
 | |
8TU9
 
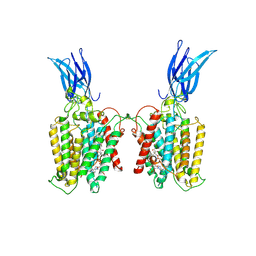 | | Cryo-EM structure of HGSNAT-acetyl-CoA complex at pH 7.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYL COENZYME *A, Enhanced green fluorescent protein,Heparan-alpha-glucosaminide N-acetyltransferase,Isoform 2 of Heparan-alpha-glucosaminide N-acetyltransferase | | Authors: | Navratna, V, Kumar, A, Mosalaganti, S. | | Deposit date: | 2023-08-15 | | Release date: | 2024-02-07 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structure of the human heparan-alpha-glucosaminide N -acetyltransferase (HGSNAT).
Elife, 13, 2024
|
|
3U0M
 
 | |
3E5V
 
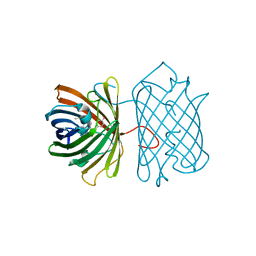 | | Crystal Structure Analysis of eqFP611 Double Mutant T122R, N143S | | Descriptor: | Red fluorescent protein eqFP611 | | Authors: | Nar, H, Nienhaus, K, Nienhaus, U, Wiedenmann, J. | | Deposit date: | 2008-08-14 | | Release date: | 2008-09-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Trans-cis isomerization is responsible for the red-shifted fluorescence in variants of the red fluorescent protein eqFP611.
J.Am.Chem.Soc., 130, 2008
|
|
3E5T
 
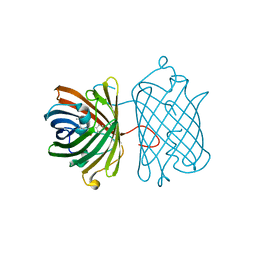 | | Crystal Structure Analysis of FP611 | | Descriptor: | Red fluorescent protein eqFP611 | | Authors: | Nar, H, Nienhaus, K, Nienhaus, U, Wiedenmann, J. | | Deposit date: | 2008-08-14 | | Release date: | 2008-09-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Trans-cis isomerization is responsible for the red-shifted fluorescence in variants of the red fluorescent protein eqFP611.
J.Am.Chem.Soc., 130, 2008
|
|
3E5W
 
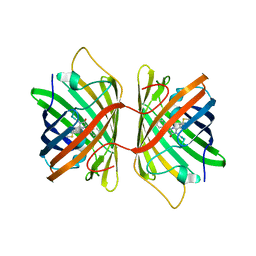 | | Crystal Structure Analysis of FP611 | | Descriptor: | Red fluorescent protein eqFP611 | | Authors: | Nienhaus, K, Nar, H, Heilker, R, Wiedenmann, J, Nienhaus, G.U. | | Deposit date: | 2008-08-14 | | Release date: | 2008-09-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Trans-cis isomerization is responsible for the red-shifted fluorescence in variants of the red fluorescent protein eqFP611.
J.Am.Chem.Soc., 130, 2008
|
|
3U0N
 
 | |
3KCS
 
 | | Crystal structure of PAmCherry1 in the dark state | | Descriptor: | PAmCherry1 protein | | Authors: | Malashkevich, V.N, Subach, F.V, Zencheck, W.D, Xiao, H, Filonov, G.S, Almo, S.C, Verkhusha, V.V. | | Deposit date: | 2009-10-21 | | Release date: | 2009-11-17 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Photoactivation mechanism of PAmCherry based on crystal structures of the protein in the dark and fluorescent states.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3KCT
 
 | | CRYSTAL STRUCTURE OF PAmCherry1 in the photoactivated state | | Descriptor: | PAmCherry1 protein | | Authors: | Malashkevich, V.N, Subach, F.V, Zencheck, W.D, Xiao, H, Filonov, G.S, Almo, S.C, Verkhusha, V.V. | | Deposit date: | 2009-10-21 | | Release date: | 2009-11-17 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Photoactivation mechanism of PAmCherry based on crystal structures of the protein in the dark and fluorescent states.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1NFQ
 
 | | Rv2002 gene product from Mycobacterium tuberculosis | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Androsterone, Putative oxidoreductase Rv2002 | | Authors: | Yang, J.K, Park, M.S, Waldo, G.S, Suh, S.W, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2002-12-15 | | Release date: | 2002-12-30 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Directed evolution approach to a structural genomics project: Rv2002 from Mycobacterium tuberculosis
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1NFF
 
 | | Crystal structure of Rv2002 gene product from Mycobacterium tuberculosis | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative oxidoreductase Rv2002 | | Authors: | Yang, J.K, Park, M.S, Waldo, G.S, Suh, S.W, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2002-12-14 | | Release date: | 2002-12-30 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Directed evolution approach to a structural genomics project: Rv2002 from Mycobacterium tuberculosis
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1NFR
 
 | | Rv2002 gene product from Mycobacterium tuberculosis | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative oxidoreductase Rv2002 | | Authors: | Yang, J.K, Park, M.S, Waldo, G.S, Suh, S.W, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2002-12-16 | | Release date: | 2002-12-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Directed evolution approach to a structural genomics project: Rv2002 from Mycobacterium tuberculosis
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1G7K
 
 | | CRYSTAL STRUCTURE OF DSRED, A RED FLUORESCENT PROTEIN FROM DISCOSOMA SP. RED | | Descriptor: | FLUORESCENT PROTEIN FP583 | | Authors: | Yarbrough, D, Wachter, R.M, Kallio, K, Matz, M.V, Remington, S.J. | | Deposit date: | 2000-11-10 | | Release date: | 2000-12-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Refined crystal structure of DsRed, a red fluorescent protein from coral, at 2.0-A resolution.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
8PN2
 
 | | CryoEM structure of Nal1 protein, allele IR64, from Oryza sativa indica cultivar | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Protein NARROW LEAF 1 | | Authors: | Huang, L.Y, Rety, S, Xi, X.G. | | Deposit date: | 2023-06-29 | | Release date: | 2024-04-17 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | The catalytic triad of rice NARROW LEAF1 involves H234.
Nat.Plants, 10, 2024
|
|
8PMM
 
 | | Structure of Nal1 protein, allele SPIKE from japonica rice, construct 31-458 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Huang, L.Y, Rety, S, Xi, X.G. | | Deposit date: | 2023-06-29 | | Release date: | 2024-04-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The catalytic triad of rice NARROW LEAF1 involves H234.
Nat.Plants, 10, 2024
|
|
8PN1
 
 | | CryoEM structure of Nal1 protein, allele SPIKE, from Oryza sativa japonica group | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Protein NARROW LEAF 1 | | Authors: | Huang, L.Y, Rety, S, Xi, X.G. | | Deposit date: | 2023-06-29 | | Release date: | 2024-04-17 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | The catalytic triad of rice NARROW LEAF1 involves H234.
Nat.Plants, 10, 2024
|
|
6ZC5
 
 | | Human Adenovirus serotype D10 FiberKnob protein | | Descriptor: | Fiber | | Authors: | Baker, A.T, Mundy, R.M, Rizkallah, P.J, Parker, A.L. | | Deposit date: | 2020-06-09 | | Release date: | 2021-06-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Development of a low-seroprevalence, alpha v beta 6 integrin-selective virotherapy based on human adenovirus type 10.
Mol Ther Oncolytics, 25, 2022
|
|
8VDR
 
 | |
3FZA
 
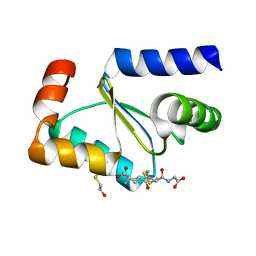 | | Crystal structure of poplar glutaredoxin S12 in complex with glutathione and beta-mercaptoethanol | | Descriptor: | BETA-MERCAPTOETHANOL, GLUTATHIONE, Glutaredoxin | | Authors: | Didierjean, C, Corbier, C, Koh, C.S, Rouhier, N, Jacquot, J.P. | | Deposit date: | 2009-01-24 | | Release date: | 2009-02-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-function relationship of the chloroplastic glutaredoxin S12 with an atypical WCSYS active site.
J.Biol.Chem., 284, 2009
|
|
3FZ9
 
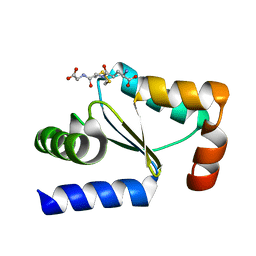 | | Crystal structure of poplar glutaredoxin S12 in complex with glutathione | | Descriptor: | GLUTATHIONE, Glutaredoxin | | Authors: | Didierjean, C, Corbier, C, Koh, C.S, Rouhier, N, Jacquot, J.P. | | Deposit date: | 2009-01-24 | | Release date: | 2009-02-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-function relationship of the chloroplastic glutaredoxin S12 with an atypical WCSYS active site.
J.Biol.Chem., 284, 2009
|
|
3IR8
 
 | | Red fluorescent protein mKeima at pH 7.0 | | Descriptor: | Large stokes shift fluorescent protein | | Authors: | Henderson, J.N, Osborn, M.F, Koon, N, Gepshtein, R, Huppert, D, Remington, S.J. | | Deposit date: | 2009-08-21 | | Release date: | 2009-09-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Excited state proton transfer in the red fluorescent protein mKeima.
J.Am.Chem.Soc., 131, 2009
|
|
