8A5D
 
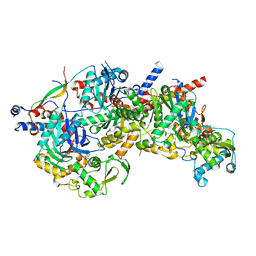 | | Structure of Arp4-Ies4-N-actin-Arp8-Ino80HSA subcomplex (A-module) of Chaetomium thermophilum INO80 | | Descriptor: | Actin, Actin related protein 4 (Arp4), Actin-related protein 8, ... | | Authors: | Kunert, F, Metzner, F.J, Eustermann, S, Jung, J, Woike, S, Schall, K, Kostrewa, D, Hopfner, K.P. | | Deposit date: | 2022-06-14 | | Release date: | 2022-12-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural mechanism of extranucleosomal DNA readout by the INO80 complex.
Sci Adv, 8, 2022
|
|
8XAY
 
 | |
7UHE
 
 | |
3N4Q
 
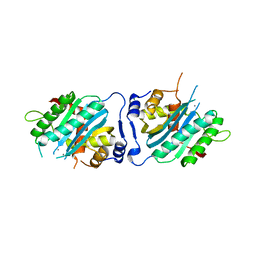 | | Human cytomegalovirus terminase nuclease domain, Mn soaked | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, TERMINASE SUBUNIT UL89 PROTEIN | | Authors: | Nadal, M, Mas, P.J, Blanco, A.G, Arnan, C, Sola, M, Hart, D.J, Coll, M. | | Deposit date: | 2010-05-22 | | Release date: | 2010-10-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure and inhibition of herpesvirus DNA packaging terminase nuclease domain.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3EBE
 
 | |
3N4P
 
 | | Human cytomegalovirus terminase nuclease domain | | Descriptor: | MAGNESIUM ION, Terminase subunit UL89 protein | | Authors: | Nadal, M, Mas, P.J, Blanco, A.G, Arnan, C, Sola, M, Hart, D.J, Coll, M. | | Deposit date: | 2010-05-22 | | Release date: | 2010-10-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure and inhibition of herpesvirus DNA packaging terminase nuclease domain.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2OBZ
 
 | |
2Y7C
 
 | | Atomic model of the Ocr-bound methylase complex from the Type I restriction-modification enzyme EcoKI (M2S1). Based on fitting into EM map 1534. | | Descriptor: | GENE 0.3 PROTEIN, TYPE I RESTRICTION ENZYME ECOKI M PROTEIN, TYPE-1 RESTRICTION ENZYME ECOKI SPECIFICITY PROTEIN | | Authors: | Kennaway, C.K, Obarska-Kosinska, A, White, J.H, Tuszynska, I, Cooper, L.P, Bujnicki, J.M, Trinick, J, Dryden, D.T.F. | | Deposit date: | 2011-01-31 | | Release date: | 2011-02-09 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (18 Å) | | Cite: | The Structure of M.Ecoki Type I DNA Methyltransferase with a DNA Mimic Antirestriction Protein.
Nucleic Acids Res., 37, 2009
|
|
5TGE
 
 | |
6OAA
 
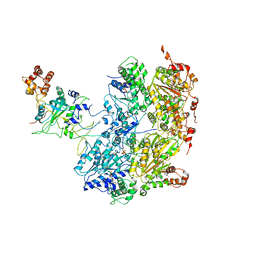 | | Cdc48-Npl4 complex processing poly-ubiquitinated substrate in the presence of ADP-BeFx, state 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Cell division control protein 48, ... | | Authors: | Twomey, E.C, Ji, Z, Wales, T.E, Bodnar, N.O, Engen, J.R, Rapoport, T.A. | | Deposit date: | 2019-03-15 | | Release date: | 2019-07-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Substrate processing by the Cdc48 ATPase complex is initiated by ubiquitin unfolding.
Science, 365, 2019
|
|
6OA9
 
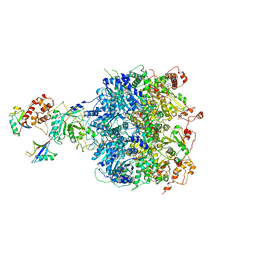 | | Cdc48-Npl4 complex processing poly-ubiquitinated substrate in the presence of ATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 48, ... | | Authors: | Twomey, E.C, Ji, Z, Wales, T.E, Bodnar, N.O, Engen, J.R, Rapoport, T.A. | | Deposit date: | 2019-03-15 | | Release date: | 2019-07-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Substrate processing by the Cdc48 ATPase complex is initiated by ubiquitin unfolding.
Science, 365, 2019
|
|
3W2Y
 
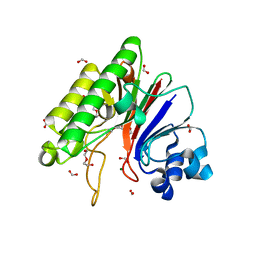 | |
3HA8
 
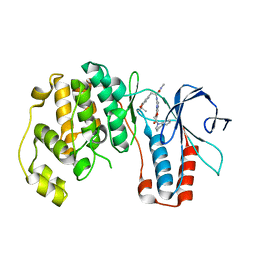 | | THE COMPLEX STRUCTURE OF THE MAP KINASE P38/Compound 14b | | Descriptor: | Mitogen-activated protein kinase 14, N~2~-{4-[6-(3,4-dihydroquinolin-1(2H)-ylcarbonyl)-1H-benzimidazol-1-yl]-6-ethoxy-1,3,5-triazin-2-yl}-3-(2,2-dimethyl-4H-1,3-benzodioxin-6-yl)-N-methyl-L-alaninamide | | Authors: | Zhao, B, Clark, M.A. | | Deposit date: | 2009-05-01 | | Release date: | 2009-08-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Design, synthesis and selection of DNA-encoded small-molecule libraries.
Nat.Chem.Biol., 5, 2009
|
|
3B2N
 
 | | Crystal structure of DNA-binding response regulator, LuxR family, from Staphylococcus aureus | | Descriptor: | SODIUM ION, Uncharacterized protein Q99UF4 | | Authors: | Malashkevich, V.N, Toro, R, Meyer, A.J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-10-18 | | Release date: | 2007-10-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal structure of DNA-binding response regulator, LuxR family, from Staphylococcus aureus.
To be Published
|
|
2PQA
 
 | | Crystal Structure of Full-length Human RPA 14/32 Heterodimer | | Descriptor: | Replication protein A 14 kDa subunit, Replication protein A 32 kDa subunit | | Authors: | Deng, X, Borgstahl, G.E. | | Deposit date: | 2007-05-01 | | Release date: | 2007-11-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the full-length human RPA14/32 complex gives insights into the mechanism of DNA binding and complex formation.
J.Mol.Biol., 374, 2007
|
|
2MJG
 
 | |
3U08
 
 | | Crystal structure of DB1963-D(CGCGAATTCGCG)2 complex at 1.25 A resolution | | Descriptor: | 4'-(5-carbamimidoyl-1H-benzimidazol-2-yl)biphenyl-4-carboxamide, 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3', MAGNESIUM ION | | Authors: | Wei, D.G, Neidle, S. | | Deposit date: | 2011-09-28 | | Release date: | 2012-09-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Small-molecule binding to the DNA minor groove is mediated by a conserved water cluster.
J.Am.Chem.Soc., 135, 2013
|
|
4O66
 
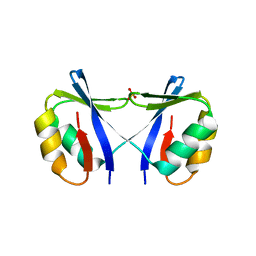 | | Crystal Structure of SMARCAL1 HARP substrate recognition domain | | Descriptor: | SODIUM ION, SULFATE ION, SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 | | Authors: | Mason, A.C, Eichman, B.F. | | Deposit date: | 2013-12-20 | | Release date: | 2014-05-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A structure-specific nucleic acid-binding domain conserved among DNA repair proteins.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7R8J
 
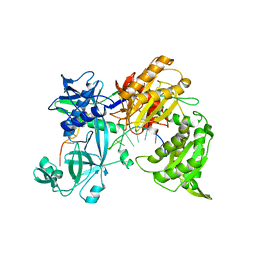 | |
5XUZ
 
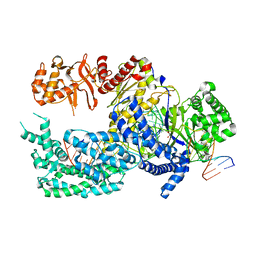 | | Crystal structure of Lachnospiraceae bacterium ND2006 Cpf1 in complex with crRNA and target DNA (CCCA PAM) | | Descriptor: | 1,2-ETHANEDIOL, DNA (29-MER), DNA (5'-D(*CP*GP*TP*CP*CP*CP*CP*CP*A)-3'), ... | | Authors: | Yamano, T, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2017-06-26 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for the Canonical and Non-canonical PAM Recognition by CRISPR-Cpf1.
Mol. Cell, 67, 2017
|
|
7MID
 
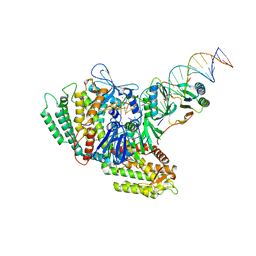 | | Sub-complex of Cas4-Cas1-Cas2 bound PAM containing DNA | | Descriptor: | CRISPR-associated endoribonuclease Cas2, CRISPR-associated exonuclease Cas4/endonuclease Cas1 fusion, DNA (33-MER), ... | | Authors: | Hu, C.Y, Ke, A.K. | | Deposit date: | 2021-04-16 | | Release date: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Mechanism for Cas4-assisted directional spacer acquisition in CRISPR-Cas.
Nature, 598, 2021
|
|
7R8G
 
 | |
7R8H
 
 | |
5FDK
 
 | |
5XUT
 
 | | Crystal structure of Lachnospiraceae bacterium ND2006 Cpf1 in complex with crRNA and target DNA (TCTA PAM) | | Descriptor: | 1,2-ETHANEDIOL, DNA (29-MER), DNA (5'-D(*CP*GP*TP*CP*CP*TP*CP*TP*A)-3'), ... | | Authors: | Yamano, T, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2017-06-26 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for the Canonical and Non-canonical PAM Recognition by CRISPR-Cpf1.
Mol. Cell, 67, 2017
|
|
