8HPZ
 
 | |
8EV5
 
 | | NlpC B3 covalently bound with E64 inhibitor fragment | | Descriptor: | Clan CA, family C40, NlpC/P60 superfamily cysteine peptidase, ... | | Authors: | Barnett, M.J, Goldstone, D.C. | | Deposit date: | 2022-10-19 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | NlpC/P60 peptidoglycan hydrolases of Trichomonas vaginalis have complementary activities that empower the protozoan to control host-protective lactobacilli.
Plos Pathog., 19, 2023
|
|
8HQ9
 
 | |
5KKG
 
 | | Crystal structure of E72A mutant of ancestral protein ancMT of ADP-dependent sugar kinases family | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCEROL, IODIDE ION, ... | | Authors: | Castro-Fernandez, V, Herrera-Morande, A, Zamora, R, Merino, F, Pereira, H.M, Brandao-Neto, J, Garratt, R, Guixe, V. | | Deposit date: | 2016-06-21 | | Release date: | 2017-07-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.608 Å) | | Cite: | Reconstructed ancestral enzymes reveal that negative selection drove the evolution of substrate specificity in ADP-dependent kinases.
J. Biol. Chem., 292, 2017
|
|
4ZTJ
 
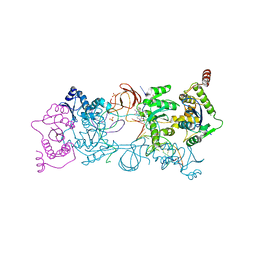 | | Crystal Structure of the Prototype Foamy Virus Intasome with a 2-Pyridinone Aminal Inhibitor | | Descriptor: | (1R,2S,5R)-8'-(3-chloro-4-fluorobenzyl)-6'-hydroxy-1-(hydroxymethyl)-2'-methyl-9',10'-dihydro-2'H-spiro[bicyclo[3.1.0]hexane-2,3'-imidazo[5,1-a][2,6]naphthyridine]-1',5',7'(8'H)-trione, DNA (5'-D(*AP*TP*TP*GP*TP*CP*AP*TP*GP*GP*AP*AP*TP*TP*TP*CP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*GP*AP*AP*AP*TP*TP*CP*CP*AP*TP*GP*AP*CP*A)-3'), ... | | Authors: | Klein, D.J, Patel, S. | | Deposit date: | 2015-05-14 | | Release date: | 2015-10-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Discovery of 2-Pyridinone Aminals: A Prodrug Strategy to Advance a Second Generation of HIV-1 Integrase Strand Transfer Inhibitors.
J.Med.Chem., 58, 2015
|
|
8ECR
 
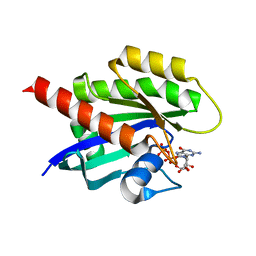 | | KRAS4b WT 1-185 bound to GDP-Mg2+ | | Descriptor: | GLYCEROL, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Betts, L, Rossman, K.L. | | Deposit date: | 2022-09-02 | | Release date: | 2023-09-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | KRAS4b WT 1-185 bound to GDP-Mg2+
To Be Published
|
|
8HQA
 
 | |
4ZRW
 
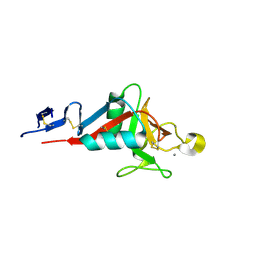 | | Structure of cow mincle complexed with trehalose | | Descriptor: | CALCIUM ION, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose, mincle protein | | Authors: | Feinberg, H, Rambaruth, N.D.S, Taylor, M.E, Drickamer, K, Weis, W.I. | | Deposit date: | 2015-05-12 | | Release date: | 2016-05-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Binding Sites for Acylated Trehalose Analogs of Glycolipid Ligands on an Extended Carbohydrate Recognition Domain of the Macrophage Receptor Mincle.
J.Biol.Chem., 291, 2016
|
|
8HM9
 
 | |
4ZVW
 
 | | Structure of apo human ALDH7A1 in space group C2 | | Descriptor: | Alpha-aminoadipic semialdehyde dehydrogenase | | Authors: | Tanner, J.J. | | Deposit date: | 2015-05-18 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of Substrate Recognition by Aldehyde Dehydrogenase 7A1.
Biochemistry, 54, 2015
|
|
4ZV7
 
 | |
8ES6
 
 | |
5KN7
 
 | | Lipid A secondary acyltransferase LpxM from Acinetobacter baumannii | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, GLYCEROL, Lipid A biosynthesis lauroyl acyltransferase, ... | | Authors: | Dovala, D.L, Hu, Q, Metzger IV, L.E. | | Deposit date: | 2016-06-27 | | Release date: | 2016-09-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure-guided enzymology of the lipid A acyltransferase LpxM reveals a dual activity mechanism.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
8HP4
 
 | | CtPDC complex | | Descriptor: | MAGNESIUM ION, Pyruvate decarboxylase, THIAMINE DIPHOSPHATE | | Authors: | Xu, H.H, Song, W. | | Deposit date: | 2022-12-11 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Protein engineering of pyruvate decarboxylase to remove the rate-limiting bottleneck in the cascade pathway of tyrosol synthesis
To Be Published
|
|
4ZXR
 
 | | Structure of Thaumatin wrapped in graphene within vacuum | | Descriptor: | GLYCEROL, L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Aller, P, Warren, A.J, Trincao, J, Evans, G. | | Deposit date: | 2015-05-20 | | Release date: | 2015-10-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | In vacuo X-ray data collection from graphene-wrapped protein crystals.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
8HP2
 
 | | CtPDC | | Descriptor: | Pyruvate decarboxylase | | Authors: | Xu, H.H, Song, W. | | Deposit date: | 2022-12-11 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Protein engineering of pyruvate decarboxylase to remove the rate-limiting bottleneck in the cascade pathway of tyrosol synthesis
To Be Published
|
|
5KKF
 
 | |
4ZW6
 
 | |
8EJO
 
 | | Crystal structure of the homeodomain of Platypus sDUX in complex with DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*GP*CP*GP*TP*AP*AP*TP*CP*TP*AP*AP*TP*CP*AP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*TP*GP*AP*TP*TP*AP*GP*AP*TP*TP*AP*CP*GP*C)-3'), ... | | Authors: | Yin, L, Shi, K, Aihara, H. | | Deposit date: | 2022-09-18 | | Release date: | 2023-09-20 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Antagonism among DUX family members evolved from an ancestral toxic single homeodomain protein.
Iscience, 26, 2023
|
|
6VKE
 
 | | Crystal Structure of Inhibitor JNJ-40012665 in Complex with Prefusion RSV F Glycoprotein | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 4-(5-chloro-2-{[1-(3,4-dimethoxyphenyl)-2-oxo-1,2-dihydro-3H-imidazo[4,5-c]pyridin-3-yl]methyl}-1H-indol-1-yl)butanenitrile, CHLORIDE ION, ... | | Authors: | McLellan, J.S. | | Deposit date: | 2020-01-20 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of 3-({5-Chloro-1-[3-(methylsulfonyl)propyl]-1H-indol-2-yl}methyl)-1-(2,2,2-trifluoroethyl)-1,3-dihydro-2H-imidazo[4,5-c]pyridin-2-one (JNJ-53718678), a Potent and Orally Bioavailable Fusion Inhibitor of Respiratory Syncytial Virus.
J.Med.Chem., 63, 2020
|
|
8EJP
 
 | | Crystal structure of the homeodomain of Platypus sDUX in complex with DNA containing 5-Bromouracil | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*GP*CP*GP*TP*AP*AP*TP*CP*TP*AP*AP*TP*CP*AP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*TP*GP*AP*TP*TP*AP*GP*AP*TP*TP*AP*CP*GP*C)-3'), ... | | Authors: | Yin, L, Shi, K, Aihara, H. | | Deposit date: | 2022-09-18 | | Release date: | 2023-09-20 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.174 Å) | | Cite: | Antagonism among DUX family members evolved from an ancestral toxic single homeodomain protein.
Iscience, 26, 2023
|
|
8EHH
 
 | | Crystal structure of the class A extended-spectrum beta-lactamase CTX-M-96 in complex with relebactam at 1.03 Angstrom resolution | | Descriptor: | (2S,5R)-1-formyl-N-(piperidin-4-yl)-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase | | Authors: | Power, P, Ghiglione, B, Bonomo, R.A, Rodriguez, M.M, Gutkind, G, Klinke, S. | | Deposit date: | 2022-09-14 | | Release date: | 2023-09-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Crystal structure of the class A extended-spectrum beta-lactamase CTX-M-96 in complex with relebactam at 1.03 Angstrom resolution.
Antimicrob.Agents Chemother., 68, 2024
|
|
4ZTI
 
 | |
8EE3
 
 | |
4ZW3
 
 | |
