1YGS
 
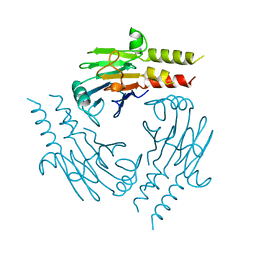 | | CRYSTAL STRUCTURE OF THE SMAD4 TUMOR SUPPRESSOR C-TERMINAL DOMAIN | | Descriptor: | SMAD4 | | Authors: | Shi, Y, Hata, A, Lo, R.S, Massague, J, Pavletich, N.P. | | Deposit date: | 1997-10-03 | | Release date: | 1998-07-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A structural basis for mutational inactivation of the tumour suppressor Smad4.
Nature, 388, 1997
|
|
3LJE
 
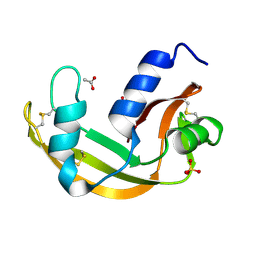 | | The X-ray structure of zebrafish RNase5 | | Descriptor: | ACETATE ION, SULFATE ION, Zebrafish RNase5 | | Authors: | Russo Krauss, I, Merlino, A, Coscia, F, Mazzarella, L, Sica, F. | | Deposit date: | 2010-01-26 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A new RNase sheds light on the RNase/angiogenin subfamily from zebrafish.
Biochem.J., 433, 2010
|
|
2Y22
 
 | | Human AlphaB-crystallin Domain (residues 67-157) | | Descriptor: | ALPHA-CRYSTALLIN B | | Authors: | Naylor, C.E, Bagneris, C, Clark, A.R, Keep, N.H, Slingsby, C. | | Deposit date: | 2010-12-13 | | Release date: | 2011-03-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Crystal Structure of R120G Disease Mutant of Human Alphab-Crystallin Domain Dimer Shows Closure of a Groove
J.Mol.Biol., 408, 2011
|
|
2XR1
 
 | | DIMERIC ARCHAEAL CLEAVAGE AND POLYADENYLATION SPECIFICITY FACTOR WITH N-TERMINAL KH DOMAINS (KH-CPSF) FROM METHANOSARCINA MAZEI | | Descriptor: | CLEAVAGE AND POLYADENYLATION SPECIFICITY FACTOR 100 KD SUBUNIT, ZINC ION | | Authors: | Mir-Montazeri, B, Ammelburg, M, Forouzan, D, Lupas, A.N, Hartmann, M.D. | | Deposit date: | 2010-09-08 | | Release date: | 2010-10-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal Structure of a Dimeric Archaeal Cleavage and Polyadenylation Specificity Factor.
J.Struct.Biol., 173, 2011
|
|
1Q52
 
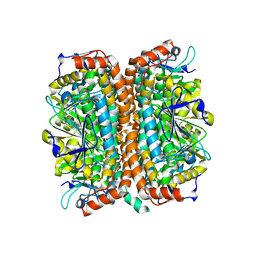 | | Crystal Structure of Mycobacterium tuberculosis MenB, a Key Enzyme in Vitamin K2 Biosynthesis | | Descriptor: | menB | | Authors: | Truglio, J.J, Theis, K, Feng, Y, Gajda, R, Machutta, C, Tonge, P.J, Kisker, C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2003-08-05 | | Release date: | 2004-01-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis MenB, a key enzyme in vitamin K2 biosynthesis.
J.Biol.Chem., 278, 2003
|
|
4OE5
 
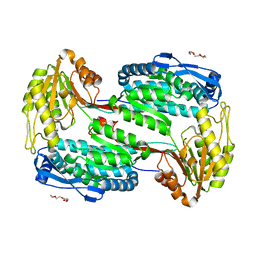 | | Structure of Human ALDH4A1 Crystallized in Space Group P21 | | Descriptor: | Delta-1-pyrroline-5-carboxylate dehydrogenase, mitochondrial, MAGNESIUM ION, ... | | Authors: | Tanner, J.J. | | Deposit date: | 2014-01-11 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Studies of Yeast Delta (1)-Pyrroline-5-carboxylate Dehydrogenase (ALDH4A1): Active Site Flexibility and Oligomeric State.
Biochemistry, 53, 2014
|
|
3KQK
 
 | |
1DCQ
 
 | | CRYSTAL STRUCTURE OF THE ARF-GAP DOMAIN AND ANKYRIN REPEATS OF PAPBETA. | | Descriptor: | PYK2-ASSOCIATED PROTEIN BETA, ZINC ION | | Authors: | Mandiyan, V, Andreev, J, Schlessinger, J, Hubbard, S.R. | | Deposit date: | 1999-11-05 | | Release date: | 1999-12-22 | | Last modified: | 2016-12-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the ARF-GAP domain and ankyrin repeats of PYK2-associated protein beta.
EMBO J., 18, 1999
|
|
6A4X
 
 | | Oxidase ChaP-H2 | | Descriptor: | Bleomycin resistance protein, FE (II) ION | | Authors: | Zhang, B, Wang, Y.S, Ge, H.M. | | Deposit date: | 2018-06-21 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Molecular Basis for the Final Oxidative Rearrangement Steps in Chartreusin Biosynthesis.
J. Am. Chem. Soc., 140, 2018
|
|
6A4Z
 
 | | Oxidase ChaP | | Descriptor: | ChaP protein, FE (II) ION | | Authors: | Zhang, B, Ge, H.M. | | Deposit date: | 2018-06-21 | | Release date: | 2018-08-29 | | Last modified: | 2018-09-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular Basis for the Final Oxidative Rearrangement Steps in Chartreusin Biosynthesis.
J. Am. Chem. Soc., 140, 2018
|
|
6I7J
 
 | | Crystal structure of monomeric FICD mutant L258D | | Descriptor: | Adenosine monophosphate-protein transferase FICD, DI(HYDROXYETHYL)ETHER, PENTAETHYLENE GLYCOL, ... | | Authors: | Perera, L.A, Yan, Y, Read, R.J, Ron, D. | | Deposit date: | 2018-11-16 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | An oligomeric state-dependent switch in the ER enzyme FICD regulates AMPylation and deAMPylation of BiP.
Embo J., 38, 2019
|
|
1DSZ
 
 | | STRUCTURE OF THE RXR/RAR DNA-BINDING DOMAIN HETERODIMER IN COMPLEX WITH THE RETINOIC ACID RESPONSE ELEMENT DR1 | | Descriptor: | DNA (5'-D(*CP*AP*GP*GP*TP*CP*AP*AP*AP*GP*GP*TP*CP*AP*G)-3'), DNA (5'-D(*CP*TP*GP*AP*CP*CP*TP*TP*TP*GP*AP*CP*CP*TP*G)-3'), RETINOIC ACID RECEPTOR ALPHA, ... | | Authors: | Rastinejad, F, Wagner, T, Zhao, Q, Khorasanizadeh, S. | | Deposit date: | 2000-01-10 | | Release date: | 2000-07-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the RXR-RAR DNA-binding complex on the retinoic acid response element DR1.
EMBO J., 19, 2000
|
|
4NDU
 
 | |
3LJD
 
 | | The X-ray structure of zebrafish RNase1 from a new crystal form at pH 4.5 | | Descriptor: | ACETATE ION, SULFATE ION, Zebrafish RNase1 | | Authors: | Russo Krauss, I, Merlino, A, Mazzarella, L, Sica, F. | | Deposit date: | 2010-01-26 | | Release date: | 2010-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | A new RNase sheds light on the RNase/angiogenin subfamily from zebrafish.
Biochem.J., 433, 2010
|
|
4NDS
 
 | |
4NSL
 
 | |
3LQY
 
 | | Crystal structure of putative isochorismatase hydrolase from Oleispira antarctica | | Descriptor: | GLYCEROL, putative isochorismatase hydrolase | | Authors: | Goral, A, Chruszcz, M, Kagan, O, Cymborowski, M, Savchenko, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-02-10 | | Release date: | 2010-03-16 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of a putative isochorismatase hydrolase from Oleispira antarctica.
J.Struct.Funct.Genom., 13, 2012
|
|
4NDT
 
 | |
2CLB
 
 | | The structure of the DPS-like protein from Sulfolobus solfataricus reveals a bacterioferritin-like di-metal binding site within a Dps- like dodecameric assembly | | Descriptor: | DPS-LIKE PROTEIN, FE (III) ION, ZINC ION | | Authors: | Gauss, G.H, Benas, P, Wiedenheft, B, Young, M, Douglas, T, Lawrence, C.M. | | Deposit date: | 2006-04-26 | | Release date: | 2006-07-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the Dps-Like Protein from Sulfolobus Solfataricus Reveals a Bacterioferritin-Like Dimetal Binding Site within a Dps-Like Dodecameric Assembly.
Biochemistry, 45, 2006
|
|
2FDO
 
 | | Crystal Structure of the Conserved Protein of Unknown Function AF2331 from Archaeoglobus fulgidus DSM 4304 Reveals a New Type of Alpha/Beta Fold | | Descriptor: | Hypothetical protein AF2331 | | Authors: | Wang, S, Kirillova, O, Chruszcz, M, Cymborowski, M.T, Skarina, T, Gorodichtchenskaia, E, Savchenko, A, Edwards, A.M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-14 | | Release date: | 2006-01-31 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of the AF2331 protein from Archaeoglobus fulgidus DSM 4304 forms an unusual interdigitated dimer with a new type of alpha + beta fold.
Protein Sci., 18, 2009
|
|
4M9P
 
 | |
2FTY
 
 | |
2FSJ
 
 | | Crystal structure of Ta0583, an archaeal actin homolog, native data | | Descriptor: | GLYCEROL, hypothetical protein Ta0583 | | Authors: | Roeben, A, Kofler, C, Nagy, I, Nickell, S, Ulrich Hartl, F, Bracher, A. | | Deposit date: | 2006-01-23 | | Release date: | 2006-04-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of an archaeal actin homolog
J.Mol.Biol., 358, 2006
|
|
2FTW
 
 | |
2FVK
 
 | |
