6OWI
 
 | | Crystal structure of MYST acetyltransferase domain in complex with inhibitor 85 | | Descriptor: | GLYCEROL, Histone acetyltransferase KAT8, N'-[(2-fluorophenyl)sulfonyl]benzohydrazide, ... | | Authors: | Hermans, S.J, Chung, M.C, Parker, M.W, Thomas, T, Baell, J.B. | | Deposit date: | 2019-05-09 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery of Acylsulfonohydrazide-Derived Inhibitors of the Lysine Acetyltransferase, KAT6A, as Potent Senescence-Inducing Anti-Cancer Agents.
J.Med.Chem., 63, 2020
|
|
4URI
 
 | | Crystal structure of chitinase-like agglutinin RobpsCRA from Robinia pseudoacacia | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHITINASE-RELATED AGGLUTININ, CHLORIDE ION, ... | | Authors: | Sulzenbacher, G, Roig-Zamboni, V, Peumans, W.J, Henrissat, B, van Damme, E.J.M, Bourne, Y. | | Deposit date: | 2014-06-30 | | Release date: | 2015-03-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Basis for Carbohydrate Binding Properties of a Plant Chitinase-Like Agglutinin with Conserved Catalytic Machinery.
J.Struct.Biol., 190, 2015
|
|
4UV6
 
 | |
7PZR
 
 | | Cryo-EM structure of POLRMT in free form. | | Descriptor: | DNA-directed RNA polymerase, mitochondrial | | Authors: | Das, H, Hallberg, B.M. | | Deposit date: | 2021-10-13 | | Release date: | 2022-11-16 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Non-coding 7S RNA inhibits transcription via mitochondrial RNA polymerase dimerization.
Cell, 185, 2022
|
|
7NP3
 
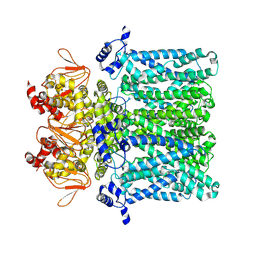 | | cAMP-free rabbit HCN4 stabilized in LMNG-CHS detergent mixture | | Descriptor: | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4,Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 | | Authors: | Giese, H.M, Chaves-Sanjuan, A, Saponaro, A, Clarke, O, Bolognesi, M, Mancia, F, Hendrickson, W.A, Thiel, G, Santoro, B, Moroni, A. | | Deposit date: | 2021-02-26 | | Release date: | 2021-08-11 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Gating movements and ion permeation in HCN4 pacemaker channels.
Mol.Cell, 81, 2021
|
|
4V31
 
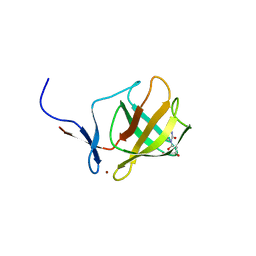 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with Deoxyuridine | | Descriptor: | 2'-DEOXYURIDINE, CEREBLON ISOFORM 4, CITRATE ANION, ... | | Authors: | Hartmann, M.D, Lupas, A.N, Hernandez Alvarez, B. | | Deposit date: | 2014-10-15 | | Release date: | 2014-12-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Thalidomide Mimics Uridine Binding to an Aromatic Cage in Cereblon.
J.Struct.Biol., 188, 2014
|
|
7SF3
 
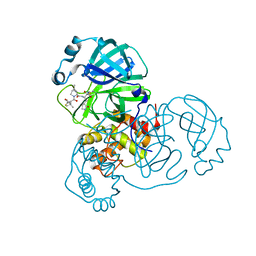 | | SARS-CoV-2 Main Protease (Mpro) in Complex with ML1006m | | Descriptor: | (1R,2S,5S)-N-{(2S,3R)-3-hydroxy-4-(methylamino)-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase, CHLORIDE ION | | Authors: | Westberg, M, Fernandez, D, Lin, M.Z. | | Deposit date: | 2021-10-02 | | Release date: | 2022-10-05 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | An orally bioavailable SARS-CoV-2 main protease inhibitor exhibits improved affinity and reduced sensitivity to mutations.
Sci Transl Med, 16, 2024
|
|
8EJL
 
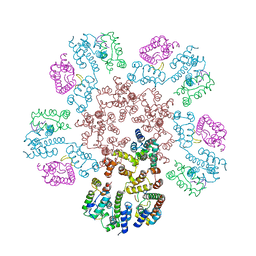 | | Structure of HIV-1 capsid declination in complex with CPSF6-FG peptide | | Descriptor: | Cleavage and polyadenylation specificity factor subunit 6, HIV-1 capsid protein | | Authors: | Pornillos, O, Ganser-Pornillos, B.K, Schirra, R.T, dos Santos, N.F.B. | | Deposit date: | 2022-09-17 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | A molecular switch modulates assembly and host factor binding of the HIV-1 capsid.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8EQP
 
 | | Crystal structure of E.coli DsbA mutant E24A/E37A/K58A | | Descriptor: | CITRATE ANION, GLYCEROL, Thiol:disulfide interchange protein DsbA | | Authors: | Wang, G, Heras, B. | | Deposit date: | 2022-10-09 | | Release date: | 2023-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Buried Water Network Modulates the Activity of the Escherichia coli Disulphide Catalyst DsbA.
Antioxidants, 12, 2023
|
|
7SET
 
 | | SARS-CoV-2 Main Protease (Mpro) in Complex with ML1000 | | Descriptor: | (1R,2S,5S)-N-{(2S,3R)-4-amino-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase, CHLORIDE ION | | Authors: | Westberg, M, Fernandez, D, Lin, M.Z. | | Deposit date: | 2021-10-01 | | Release date: | 2022-10-05 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | An orally bioavailable SARS-CoV-2 main protease inhibitor exhibits improved affinity and reduced sensitivity to mutations.
Sci Transl Med, 16, 2024
|
|
4WGI
 
 | | A Single Diastereomer of a Macrolactam Core Binds Specifically to Myeloid Cell Leukemia 1 (MCL1) | | Descriptor: | (2S)-2-[(2S,3R)-10-{[(4-fluorophenyl)sulfonyl]amino}-3-methyl-2-[(methyl{[4-(trifluoromethyl)phenyl]carbamoyl}amino)methyl]-6-oxo-3,4-dihydro-2H-1,5-benzoxazocin-5(6H)-yl]propanoic acid, FORMIC ACID, MAGNESIUM ION, ... | | Authors: | Clifton, M.C, Fairman, J.W, Fang, C, D'Souza, B, Fulroth, B, Leed, A, McCarren, P, Wang, L, Wang, Y, Kaushik, V, Palmer, M, Wei, G, Golub, T.R, Hubbard, B.K, Serrano-Wu, M.H. | | Deposit date: | 2014-09-18 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Single Diastereomer of a Macrolactam Core Binds Specifically to Myeloid Cell Leukemia 1 (MCL1).
Acs Med.Chem.Lett., 5, 2014
|
|
8EPG
 
 | | Engineering Crystals with Tunable Symmetries from 14- or 16-Base-Long DNA Strands | | Descriptor: | DNA (5'-D(*CP*GP*AP*CP*G)-3'), DNA (5'-D(P*CP*GP*TP*GP*GP*C)-3'), DNA (5'-D(P*GP*CP*CP*GP*C)-3') | | Authors: | Zhang, C, Zhao, J, Lu, B, Sha, R, Seeman, N.C, Noinaj, N, Mao, C. | | Deposit date: | 2022-10-05 | | Release date: | 2023-03-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Engineering DNA Crystals toward Studying DNA-Guest Molecule Interactions.
J.Am.Chem.Soc., 145, 2023
|
|
4WJT
 
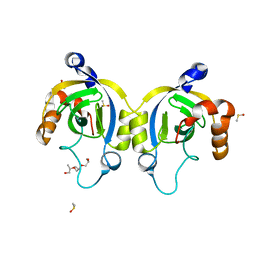 | | Stationary Phase Survival Protein YuiC from B.subtilis complexed with NAG | | Descriptor: | (2S)-2-{[(2S)-2-{[(2R)-2-hydroxypropyl]oxy}propyl]oxy}propan-1-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Quay, D.H.X, Cole, A.R, Cryar, A, Thalassinos, K, Williams, M.A, Bhakta, S, Keep, N.H. | | Deposit date: | 2014-10-01 | | Release date: | 2015-07-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Structure of the stationary phase survival protein YuiC from B.subtilis.
Bmc Struct.Biol., 15, 2015
|
|
8EPI
 
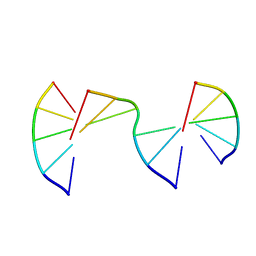 | | Engineering Crystals with Tunable Symmetries from 14- or 16-Base-Long DNA Strands | | Descriptor: | DNA (5'-D(*CP*GP*AP*CP*G)-3'), DNA (5'-D(P*CP*GP*TP*GP*GP*A)-3'), DNA (5'-D(P*TP*CP*CP*TP*A)-3') | | Authors: | Zhang, C, Zhao, J, Lu, B, Sha, R, Seeman, N.C, Noinaj, N, Mao, C. | | Deposit date: | 2022-10-05 | | Release date: | 2023-03-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Engineering DNA Crystals toward Studying DNA-Guest Molecule Interactions.
J.Am.Chem.Soc., 145, 2023
|
|
8EPD
 
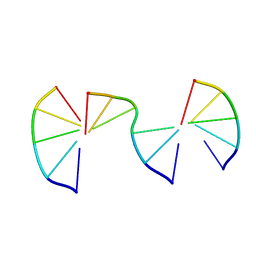 | | Engineering Crystals with Tunable Symmetries from 14- or 16-Base-Long DNA Strands | | Descriptor: | DNA (5'-D(*CP*GP*AP*CP*G)-3'), DNA (5'-D(P*CP*GP*TP*TP*CP*C)-3'), DNA (5'-D(P*GP*GP*AP*GP*C)-3') | | Authors: | Zhang, C, Zhao, J, Lu, B, Sha, R, Seeman, N.C, Noinaj, N, Mao, C. | | Deposit date: | 2022-10-05 | | Release date: | 2023-03-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Engineering DNA Crystals toward Studying DNA-Guest Molecule Interactions.
J.Am.Chem.Soc., 145, 2023
|
|
6XA0
 
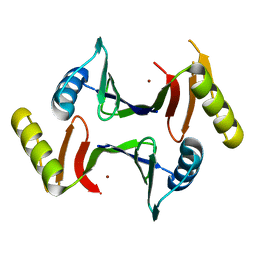 | | Crystal structure of C-As lyase with mutation K105R with Ni(II) | | Descriptor: | Glyoxalase/bleomycin resistance protein/dioxygenase, NICKEL (II) ION | | Authors: | Venkadesh, S, Yoshinaga, M, Kandavelu, P, Sankaran, B, Rosen, B.P. | | Deposit date: | 2020-06-03 | | Release date: | 2021-06-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The ArsI C-As lyase: Elucidating the catalytic mechanism of degradation of organoarsenicals.
J.Inorg.Biochem., 232, 2022
|
|
6XCK
 
 | | Crystal structure of C-As lyase with mutation K105E | | Descriptor: | Glyoxalase/bleomycin resistance protein/dioxygenase | | Authors: | Venkadesh, S, Yoshinaga, M, Kandavelu, P, Sankaran, B, Rosen, B.P. | | Deposit date: | 2020-06-08 | | Release date: | 2021-06-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | The ArsI C-As lyase: Elucidating the catalytic mechanism of degradation of organoarsenicals.
J.Inorg.Biochem., 232, 2022
|
|
8EP8
 
 | | Engineering Crystals with Tunable Symmetries from 14- or 16-Base-Long DNA Strands | | Descriptor: | DNA (5'-D(*CP*GP*AP*CP*G)-3'), DNA (5'-D(P*CP*GP*TP*GP*GP*A)-3'), DNA (5'-D(P*TP*CP*CP*GP*C)-3') | | Authors: | Zhang, C, Zhao, J, Lu, B, Sha, R, Seeman, N.C, Noinaj, N, Mao, C. | | Deposit date: | 2022-10-05 | | Release date: | 2023-03-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Engineering DNA Crystals toward Studying DNA-Guest Molecule Interactions.
J.Am.Chem.Soc., 145, 2023
|
|
4WLN
 
 | | Crystal structure of apo MDH2 | | Descriptor: | Malate dehydrogenase, mitochondrial, PHOSPHATE ION | | Authors: | Eo, Y.M, Han, B.G, Ahn, H.C. | | Deposit date: | 2014-10-07 | | Release date: | 2015-11-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structure of apo MDH2
To Be Published
|
|
7SF1
 
 | | SARS-CoV-2 Main Protease (Mpro) in Complex with ML1001 | | Descriptor: | (1R,2S,5S)-N-{(2S,3R)-4-amino-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-3-[N-(3,3-dimethylbutanoyl)-3-methyl-L-valyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Westberg, M, Fernandez, D, Lin, M.Z. | | Deposit date: | 2021-10-02 | | Release date: | 2022-10-05 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | An orally bioavailable SARS-CoV-2 main protease inhibitor exhibits improved affinity and reduced sensitivity to mutations.
Sci Transl Med, 16, 2024
|
|
8SW7
 
 | | BG505 Boost2 SOSIP.664 in complex with NHP polyclonal antibody FP1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BG505 Boost 2 gp120, BG505 Boost 2 gp41, ... | | Authors: | Pratap, P.P, Antansijevic, A, Ozorowski, G, Ward, A.B. | | Deposit date: | 2023-05-17 | | Release date: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Focusing antibody responses to the fusion peptide in rhesus macaques.
Biorxiv, 2023
|
|
5HIX
 
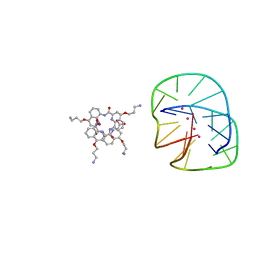 | | Cocrystal structure of an anti-parallel DNA G-quadruplex and a tetra-Quinoline Foldamer | | Descriptor: | 4-(3-aminopropoxy)-8-({[4-(3-aminopropoxy)-8-({[4-(3-aminopropoxy)-8-({[4-(3-aminopropoxy)-8-nitroquinolin-2-yl]carbonyl}amino)quinolin-2-yl]carbonyl}amino)quinolin-2-yl]carbonyl}amino)quinoline-2-carboxylic acid, Dimeric G-quadruplex, POTASSIUM ION | | Authors: | Mandal, P.K, Baptiste, B, Langlois d'Estaintot, B, Kauffmann, B, Huc, I. | | Deposit date: | 2016-01-12 | | Release date: | 2016-11-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Multivalent Interactions between an Aromatic Helical Foldamer and a DNA G-Quadruplex in the Solid State.
Chembiochem, 17, 2016
|
|
4WJG
 
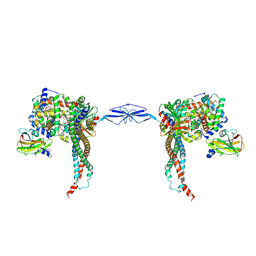 | | Structure of T. brucei haptoglobin-hemoglobin receptor binding to human haptoglobin-hemoglobin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Haptoglobin, ... | | Authors: | Stoedkilde, K, Torvund-Jensen, M, Moestrup, S.K, Andersen, C.B.F. | | Deposit date: | 2014-09-30 | | Release date: | 2014-11-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for trypanosomal haem acquisition and susceptibility to the host innate immune system.
Nat Commun, 5, 2014
|
|
6UFE
 
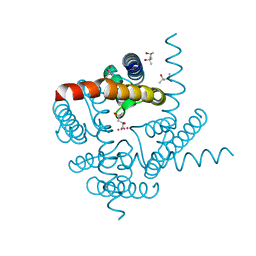 | | The structure of a potassium selective ion channel at atomic resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, POTASSIUM ION, Transporter | | Authors: | Langan, P.S, Vandavasi, V.G, Sullivan, B, Afonine, P.V, Weiss, K.L. | | Deposit date: | 2019-09-24 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The structure of a potassium-selective ion channel reveals a hydrophobic gate regulating ion permeation.
Iucrj, 7, 2020
|
|
8F76
 
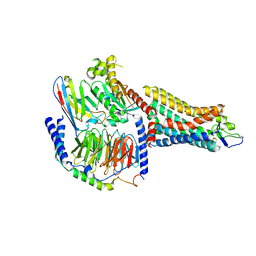 | | Human olfactory receptor OR51E2 bound to propionate in complex with miniGs399 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Billesboelle, C.B, Manglik, A. | | Deposit date: | 2022-11-18 | | Release date: | 2023-03-22 | | Last modified: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of odorant recognition by a human odorant receptor.
Nature, 615, 2023
|
|
