9B5N
 
 | |
9B5M
 
 | |
9B5L
 
 | |
9B5K
 
 | |
9B5J
 
 | |
9B5I
 
 | |
9B5H
 
 | |
9B5G
 
 | |
9B5F
 
 | |
9B5E
 
 | |
9B5D
 
 | |
9B5C
 
 | |
9B5B
 
 | |
9B5A
 
 | |
9B59
 
 | |
9B58
 
 | |
9B57
 
 | |
9B56
 
 | |
9B55
 
 | |
9B4H
 
 | | Chlamydomonas reinhardtii mastigoneme filament | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C-type lectin domain-containing protein, Tyrosine-protein kinase ephrin type A/B receptor-like domain-containing protein, ... | | Authors: | Dai, J, Ma, M, Zhang, R, Brown, A. | | Deposit date: | 2024-03-20 | | Release date: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mastigoneme structure reveals insights into the O-linked glycosylation code of native hydroxyproline-rich helices.
Cell, 2024
|
|
9B4G
 
 | | Structure of inhibitor-bound human PSS1 | | Descriptor: | (2R)-3-{[(S)-hydroxy{[(1R,2R,3R,4R,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dihexadecanoate, (7P)-7-[(4S)-4-{4-[3,5-bis(trifluoromethyl)phenoxy]phenyl}-5-(2,2-difluoropropyl)-6-oxo-1,4,5,6-tetrahydropyrrolo[3,4-c]pyrazol-3-yl]-1,3-benzoxazol-2(3H)-one, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, ... | | Authors: | Long, T, Li, X. | | Deposit date: | 2024-03-20 | | Release date: | 2024-09-04 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Molecular insights into human phosphatidylserine synthase 1 reveal its inhibition promotes LDL uptake.
Cell, 2024
|
|
9B4F
 
 | | Structure of human PSS1-P269S | | Descriptor: | CALCIUM ION, Phosphatidylserine synthase 1 | | Authors: | Long, T, Li, X. | | Deposit date: | 2024-03-20 | | Release date: | 2024-09-04 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Molecular insights into human phosphatidylserine synthase 1 reveal its inhibition promotes LDL uptake.
Cell, 2024
|
|
9B4E
 
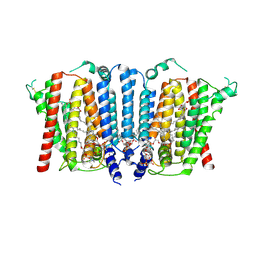 | | Structure of wild type human PSS1 | | Descriptor: | (2R)-3-{[(S)-hydroxy{[(1R,2R,3R,4R,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dihexadecanoate, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, CALCIUM ION, ... | | Authors: | Long, T, Li, X. | | Deposit date: | 2024-03-20 | | Release date: | 2024-09-04 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Molecular insights into human phosphatidylserine synthase 1 reveal its inhibition promotes LDL uptake.
Cell, 2024
|
|
9B44
 
 | |
9B3R
 
 | | The structure of human cardiac F-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha cardiac muscle 1, ... | | Authors: | Doran, M.H, Sousa, D, Rynkiewicz, M.J, Lehman, W, Cammarato, A. | | Deposit date: | 2024-03-20 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of human cardiac actin
To Be Published
|
|
