8SAP
 
 | |
8SAO
 
 | |
1H8G
 
 | | C-terminal domain of the major autolysin (C-LytA) from Streptococcus pneumoniae | | Descriptor: | CHOLINE ION, MAJOR AUTOLYSIN | | Authors: | Fernandez-Tornero, C, Garcia, E, Lopez, R, Gimenez-Gallego, G, Romero, A. | | Deposit date: | 2001-02-06 | | Release date: | 2002-01-31 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A Novel Solenoid Fold in the Cell Wall Anchoring Domain of the Pneumococcal Virulence Factor Lyta
Nat.Struct.Biol., 8, 2001
|
|
1HCX
 
 | | Choline binding domain of the major autolysin (C-LytA) from Streptococcus pneumoniae | | Descriptor: | 2,2':6',2''-TERPYRIDINE PLATINUM(II) Chloride, CHOLINE ION, DECYLAMINE-N,N-DIMETHYL-N-OXIDE, ... | | Authors: | Fernandez-Tornero, C, Lopez, R, Garcia, E, Gimenez-Gallego, G, Romero, A. | | Deposit date: | 2001-05-10 | | Release date: | 2001-11-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Novel Solenoid Fold in the Cell Wall Anchoring Domain of the Pneumococcal Virulence Factor Lyta
Nat.Struct.Biol., 8, 2001
|
|
3SLJ
 
 | | Pre-cleavage Structure of the Autotransporter EspP - N1023A mutant | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, Serine protease espP | | Authors: | Barnard, T.B, Noinaj, N, Easley, N.C, Kuszak, A.J, Buchanan, S.K. | | Deposit date: | 2011-06-24 | | Release date: | 2011-11-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.481 Å) | | Cite: | Molecular basis for the activation of a catalytic asparagine residue in a self-cleaving bacterial autotransporter.
J.Mol.Biol., 415, 2012
|
|
1ORM
 
 | |
3SLT
 
 | | Pre-cleavage Structure of the Autotransporter EspP - N1023S Mutant | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, Serine protease espP | | Authors: | Barnard, T.B, Noinaj, N, Easley, N.C, Kuszak, A.J, Buchanan, S.K. | | Deposit date: | 2011-06-25 | | Release date: | 2011-11-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Molecular basis for the activation of a catalytic asparagine residue in a self-cleaving bacterial autotransporter.
J.Mol.Biol., 415, 2012
|
|
3SLO
 
 | | Pre-cleavage Structure of the Autotransporter EspP - N1023D mutant | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, Serine protease espP | | Authors: | Barnard, T.B, Noinaj, N, Easley, N.C, Kuszak, A.J, Buchanan, S.K. | | Deposit date: | 2011-06-24 | | Release date: | 2011-11-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Molecular basis for the activation of a catalytic asparagine residue in a self-cleaving bacterial autotransporter.
J.Mol.Biol., 415, 2012
|
|
3TGK
 
 | | TRYPSINOGEN MUTANT D194N AND DELETION OF ILE 16-VAL 17 COMPLEXED WITH BOVINE PANCREATIC TRYPSIN INHIBITOR (BPTI) | | Descriptor: | CALCIUM ION, PANCREATIC TRYPSIN INHIBITOR, SULFATE ION, ... | | Authors: | Pasternak, A, White, A, Jeffery, C.J, Medina, N, Cahoon, M, Ringe, D, Hedstrom, L. | | Deposit date: | 1998-07-19 | | Release date: | 2001-07-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The energetic cost of induced fit catalysis: Crystal structures of trypsinogen mutants with enhanced activity and inhibitor affinity.
Protein Sci., 10, 2001
|
|
6CFF
 
 | | Stimulator of Interferon Genes Human | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, Stimulator of interferon genes protein | | Authors: | Fernandez, D, Li, L, Ergun, S.L. | | Deposit date: | 2018-02-14 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.396 Å) | | Cite: | STING Polymer Structure Reveals Mechanisms for Activation, Hyperactivation, and Inhibition.
Cell, 178, 2019
|
|
6CY7
 
 | | Human Stimulator of Interferon Genes | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, GLYCEROL, IMIDAZOLE, ... | | Authors: | Fernandez, D, Li, L, Ergun, S.L. | | Deposit date: | 2018-04-04 | | Release date: | 2019-03-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | STING Polymer Structure Reveals Mechanisms for Activation, Hyperactivation, and Inhibition.
Cell, 178, 2019
|
|
368D
 
 | | STRUCTURAL VARIABILITY OF A-DNA IN CRYSTALS OF THE OCTAMER D(PCPCPCPGPCPGPGPG) | | Descriptor: | DNA (5'-D(P*CP*CP*CP*GP*CP*GP*GP*G)-3') | | Authors: | Fernandez, L.G, Subirana, J.A, Verdaguer, N, Pyshnyi, D, Campos, L, Malinina, L. | | Deposit date: | 1997-12-19 | | Release date: | 1998-07-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural variability of A-DNA in crystals of the octamer d(pCpCpCpGpCpGpGpG)
J.Biomol.Struct.Dyn., 15, 1997
|
|
2BOI
 
 | | 1.1A Structure of Chromobacterium Violaceum Lectin CV2L in Complex with alpha-methyl-fucoside | | Descriptor: | CALCIUM ION, CV-IIL LECTIN, methyl alpha-L-fucopyranoside | | Authors: | Pokorna, M, Cioci, G, Perret, S, Rebuffet, E, Adam, J, Gilboa-Garber, N, Mitchell, E.P, Imberty, A, Wimmerova, M. | | Deposit date: | 2005-04-12 | | Release date: | 2006-05-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Unusual Entropy Driven Affinity of Chromobacter Violaceum Lectin Cv-Iil Towards Fucose and Mannose
Biochemistry, 45, 2006
|
|
371D
 
 | | STRUCTURAL VARIABILITY OF A-DNA IN CRYSTALS OF THE OCTAMER D(PCPCPCPGPCPGPGPG) | | Descriptor: | DNA (5'-D(P*CP*CP*CP*GP*CP*GP*GP*G)-3') | | Authors: | Fernandez, L.G, Subirana, J.A, Verdaguer, N, Pyshnyi, D, Campos, L. | | Deposit date: | 1997-12-19 | | Release date: | 1998-07-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural variability of A-DNA in crystals of the octamer d(pCpCpCpGpCpGpGpG)
J.Biomol.Struct.Dyn., 15, 1997
|
|
370D
 
 | | STRUCTURAL VARIABILITY OF A-DNA IN CRYSTALS OF THE OCTAMER D(PCPCPCPGPCPGPGPG) | | Descriptor: | DNA (5'-D(P*CP*CP*CP*GP*CP*GP*GP*G)-3'), MAGNESIUM ION | | Authors: | Fernandez, L.G, Subirana, J.A, Verdaguer, N, Pyshnyi, D, Campos, L. | | Deposit date: | 1997-12-19 | | Release date: | 1998-07-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural variability of A-DNA in crystals of the octamer d(pCpCpCpGpCpGpGpG)
J.Biomol.Struct.Dyn., 15, 1997
|
|
369D
 
 | | STRUCTURAL VARIABILITY OF A-DNA IN CRYSTALS OF THE OCTAMER D(PCPCPCPGPCPGPGPG) | | Descriptor: | DNA (5'-D(P*CP*CP*CP*GP*CP*GP*GP*G)-3') | | Authors: | Fernandez, L.G, Subirana, J.A, Verdaguer, N, Pyshnyi, D, Campos, L. | | Deposit date: | 1997-12-19 | | Release date: | 1998-07-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural variability of A-DNA in crystals of the octamer d(pCpCpCpGpCpGpGpG)
J.Biomol.Struct.Dyn., 15, 1997
|
|
2BV4
 
 | | 1.0A Structure of Chromobacterium Violaceum Lectin in Complex with alpha-methyl-mannoside | | Descriptor: | CALCIUM ION, LECTIN CV-IIL, methyl alpha-D-mannopyranoside | | Authors: | Pokorna, M, Cioci, G, Perret, S, Rebuffet, E, Adam, J, Gilboa-Garber, N, Mitchell, E.P, Imberty, A, Wimmerova, M. | | Deposit date: | 2005-06-22 | | Release date: | 2006-05-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Unusual Entropy Driven Affinity of Chromobacterium Violaceum Lectin Cv-Iil Towards Fucose and Mannose
Biochemistry, 45, 2006
|
|
3TGI
 
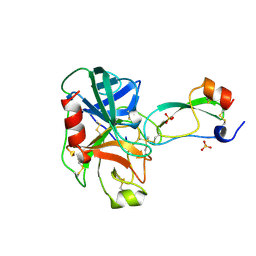 | | WILD-TYPE RAT ANIONIC TRYPSIN COMPLEXED WITH BOVINE PANCREATIC TRYPSIN INHIBITOR (BPTI) | | Descriptor: | BOVINE PANCREATIC TRYPSIN INHIBITOR, CALCIUM ION, SULFATE ION, ... | | Authors: | Pasternak, A, Ringe, D, Hedstrom, L. | | Deposit date: | 1998-07-15 | | Release date: | 1998-12-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Comparison of Anionic and Cationic Trypsinogens: The Anionic Activation Domain is More Flexible in Solution and Differs in its Mode of Bpti Binding in the Crystal Structure
Protein Sci., 8, 1999
|
|
3TGJ
 
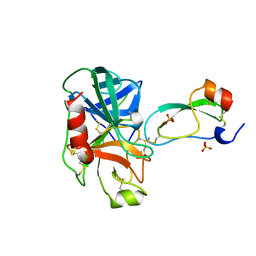 | | S195A TRYPSINOGEN COMPLEXED WITH BOVINE PANCREATIC TRYPSIN INHIBITOR (BPTI) | | Descriptor: | BOVINE PANCREATIC TRYPSIN INHIBITOR, CALCIUM ION, SULFATE ION, ... | | Authors: | Pasternak, A, Ringe, D, Hedstrom, L. | | Deposit date: | 1998-07-16 | | Release date: | 1998-12-23 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Comparison of Anionic and Cationic Trypsinogens: The Anionic Activation Domain is More Flexible in Solution and Differs in its Mode of Bpti Binding in the Crystal Structure
Protein Sci., 8, 1999
|
|
372D
 
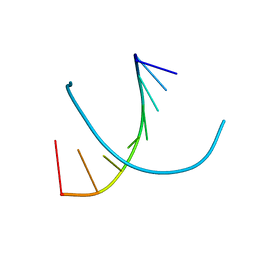 | | STRUCTURAL VARIABILITY OF A-DNA IN CRYSTALS OF THE OCTAMER D(PCPCPCPGPCPGPGPG) | | Descriptor: | DNA (5'-D(P*CP*CP*CP*GP*CP*GP*GP*G)-3') | | Authors: | Fernandez, L.G, Subirana, J.A, Verdaguer, N, Pyshnyi, D, Campos, L. | | Deposit date: | 1997-12-19 | | Release date: | 1998-07-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural variability of A-DNA in crystals of the octamer d(pCpCpCpGpCpGpGpG)
J.Biomol.Struct.Dyn., 15, 1997
|
|
6DNK
 
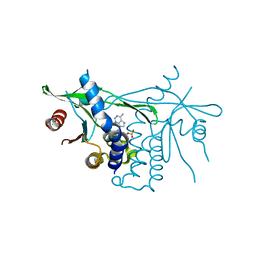 | | Human Stimulator of Interferon Genes | | Descriptor: | Stimulator of interferon genes protein, cGAMP | | Authors: | Fernandez, D, Li, L, Ergun, S.L. | | Deposit date: | 2018-06-06 | | Release date: | 2019-03-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | STING Polymer Structure Reveals Mechanisms for Activation, Hyperactivation, and Inhibition.
Cell, 178, 2019
|
|
1CM9
 
 | |
1F5R
 
 | | RAT TRYPSINOGEN MUTANT COMPLEXED WITH BOVINE PANCREATIC TRYPSIN INHIBITOR | | Descriptor: | CALCIUM ION, PANCREATIC TRYPSIN INHIBITOR, TRYPSIN II, ... | | Authors: | Pasternak, A, White, A, Cahoon, M, Ringe, D, Hedstrom, L. | | Deposit date: | 2000-06-15 | | Release date: | 2001-07-04 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The energetic cost of induced fit catalysis: Crystal structures of trypsinogen mutants with enhanced activity and inhibitor affinity.
Protein Sci., 10, 2001
|
|
4BL1
 
 | | Crystal structure of unphosphorylated Maternal Embryonic Leucine zipper Kinase (MELK) in complex with AMP-PNP | | Descriptor: | MATERNAL EMBRYONIC LEUCINE ZIPPER KINASE, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Canevari, G, Re-Depaolini, S, Cucchi, U, Forte, B, Carpinelli, P, Bertrand, J.A. | | Deposit date: | 2013-04-30 | | Release date: | 2013-05-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Unphosphorylated Maternal Embryonic Leucine Zipper Kinase
To be Published
|
|
6E5M
 
 | | Crystallographic structure of the cyclic nonapeptide derived from the BTCI inhibitor bound to beta-trypsin in space group P 32 2 1 | | Descriptor: | 9MER-PEPTIDE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Fernandes, J.C, Valadares, N.F, Freitas, S.M, Barbosa, J.A.R.G. | | Deposit date: | 2018-07-20 | | Release date: | 2019-03-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.612 Å) | | Cite: | Crystallographic structure of a complex between trypsin and a nonapeptide derived from a Bowman-Birk inhibitor found in Vigna unguiculata seeds.
Arch. Biochem. Biophys., 665, 2019
|
|
