9XIM
 
 | |
9RLR
 
 | | PARP15 catalytic domain in complex with OUL499 | | Descriptor: | 6-[(3-chlorophenyl)methoxy]-2,3-dihydrophthalazine-1,4-dione, DIMETHYL SULFOXIDE, Protein mono-ADP-ribosyltransferase PARP15 | | Authors: | Alaviuhkola, J, Lehtio, L. | | Deposit date: | 2025-06-17 | | Release date: | 2025-09-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Optimization of 2,3-dihydrophthalazine-1,4-dione PARP inhibitor scaffold for nanomolar potency and specificity towards human PARP10.
Eur.J.Med.Chem., 300, 2025
|
|
9V7W
 
 | |
9RLO
 
 | | PARP15 catalytic domain in complex with OUL250 | | Descriptor: | 4-[[1,4-bis(oxidanylidene)-2,3-dihydrophthalazin-6-yl]oxy]benzamide, DIMETHYL SULFOXIDE, Protein mono-ADP-ribosyltransferase PARP15 | | Authors: | Alaviuhkola, J, Maksimainen, M.M, Lehtio, L. | | Deposit date: | 2025-06-17 | | Release date: | 2025-09-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Optimization of 2,3-dihydrophthalazine-1,4-dione PARP inhibitor scaffold for nanomolar potency and specificity towards human PARP10.
Eur.J.Med.Chem., 300, 2025
|
|
9RLP
 
 | | PARP15 catalytic domain in complex with OUL310 | | Descriptor: | 6-[(3-fluorophenyl)methoxy]-2,3-dihydrophthalazine-1,4-dione, DIMETHYL SULFOXIDE, Protein mono-ADP-ribosyltransferase PARP15 | | Authors: | Alaviuhkola, J, Maksimainen, M.M, Lehtio, L. | | Deposit date: | 2025-06-17 | | Release date: | 2025-09-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Optimization of 2,3-dihydrophthalazine-1,4-dione PARP inhibitor scaffold for nanomolar potency and specificity towards human PARP10.
Eur.J.Med.Chem., 300, 2025
|
|
9PGS
 
 | | HIV Capsid Hexamer bound to Compound 6 | | Descriptor: | 3,5-difluoro-Nalpha-[(5-hydroxy-1H-indol-3-yl)acetyl]-N-(4-methoxyphenyl)-N-methyl-L-phenylalaninamide, HIV-1 capsid | | Authors: | Somoza, J.R, Anderson, R.L, Villasenor, A.G, Ferrao, R.D. | | Deposit date: | 2025-07-08 | | Release date: | 2025-10-08 | | Last modified: | 2025-10-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of Lenacapavir: First-in-Class Twice-Yearly Capsid Inhibitor for HIV-1 Treatment and Pre-exposure Prophylaxis.
J.Med.Chem., 2025
|
|
9O11
 
 | |
9O13
 
 | |
9O12
 
 | |
9O10
 
 | |
9QEK
 
 | | Structure of Human ROS1 Kinase Domain Harboring the G2032R Solvent-front Mutation in Complex with Zidesamtinib (NVL-520) | | Descriptor: | 1,2-ETHANEDIOL, Proto-oncogene tyrosine-protein kinase ROS, Zidesamtinib | | Authors: | Tangpeerachaikul, A, Mente, S, Magrino, J, Gu, F, Horan, J.C, Pelish, H.E. | | Deposit date: | 2025-03-10 | | Release date: | 2025-04-30 | | Last modified: | 2025-07-16 | | Method: | X-RAY DIFFRACTION (2.205 Å) | | Cite: | Zidesamtinib Selective Targeting of Diverse ROS1 Drug-Resistant Mutations.
Mol.Cancer Ther., 24, 2025
|
|
9P8Z
 
 | |
9QT6
 
 | | Crystal structure of HPK1 T165E/S171E in complex with pyrazine carboxamide inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 3-[(1,3-dimethylpyrazol-4-yl)amino]-5-(methylamino)-6-(3-methylimidazo[4,5-c]pyridin-7-yl)pyrazine-2-carboxamide, Mitogen-activated protein kinase kinase kinase kinase 1 | | Authors: | Schimpl, M. | | Deposit date: | 2025-04-08 | | Release date: | 2025-05-28 | | Last modified: | 2025-09-03 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Synthetic chemistry enabling the discovery and development of a series of pyrazoles as HPK1 inhibitors.
Rsc Med Chem, 16, 2025
|
|
9RLS
 
 | | PARP10 catalytic domain in complex with OUL499 | | Descriptor: | 6-[(3-chlorophenyl)methoxy]-2,3-dihydrophthalazine-1,4-dione, Poly [ADP-ribose] polymerase 10 | | Authors: | Alaviuhkola, J, Lehtio, L. | | Deposit date: | 2025-06-17 | | Release date: | 2025-09-17 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Optimization of 2,3-dihydrophthalazine-1,4-dione PARP inhibitor scaffold for nanomolar potency and specificity towards human PARP10.
Eur.J.Med.Chem., 300, 2025
|
|
9VXS
 
 | |
9R5T
 
 | | NSP14 IN COMPLEX WITH LIGAND TDI-016037-NX-1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Guanine-N7 methyltransferase nsp14, ... | | Authors: | Steinbacher, S, Meyer, C. | | Deposit date: | 2025-05-09 | | Release date: | 2025-08-27 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Discovery of Novel Isofunctional SARS-CoV‐2 NSP14 RNA Cap Methyltransferase Inhibitors by Structure-Based Virtual Screening.
Acs Med.Chem.Lett., 16, 2025
|
|
9OH7
 
 | | M13F/H117A/M121H Azurin with Cu(II), pH 7.4 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Azurin, COPPER (II) ION | | Authors: | Van Stappen, C, Lu, Y. | | Deposit date: | 2025-05-02 | | Release date: | 2025-07-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Beyond Blue: Systematic Modulation of Electronic Structure and Redox Properties of Type 1 Copper in Azurin.
J.Am.Chem.Soc., 147, 2025
|
|
9O90
 
 | |
6P2I
 
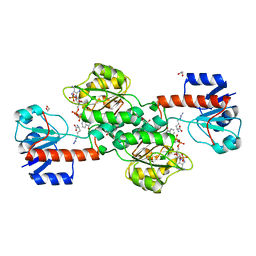 | | Acyclic imino acid reductase (Bsp5) in complex with NADPH and D-Arg | | Descriptor: | 1,2-ETHANEDIOL, D-ARGININE, Glycerate dehydrogenase, ... | | Authors: | Guo, J, Higgins, M.A, Daniel-Ivad, P, Ryan, K.S. | | Deposit date: | 2019-05-21 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | An Asymmetric Reductase That Intercepts Acyclic Imino Acids Producedin Situby a Partner Oxidase.
J.Am.Chem.Soc., 141, 2019
|
|
9RLQ
 
 | | PARP15 catalytic domain in complex with OUL312 | | Descriptor: | 6-[2-(2-fluorophenyl)ethoxy]-2,3-dihydrophthalazine-1,4-dione, DIMETHYL SULFOXIDE, Protein mono-ADP-ribosyltransferase PARP15 | | Authors: | Alaviuhkola, J, Maksimainen, M.M, Lehtio, L. | | Deposit date: | 2025-06-17 | | Release date: | 2025-09-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Optimization of 2,3-dihydrophthalazine-1,4-dione PARP inhibitor scaffold for nanomolar potency and specificity towards human PARP10.
Eur.J.Med.Chem., 300, 2025
|
|
9NPG
 
 | | X-ray crystal structure of recombinant Can f 1-C100S in complex with human IgE mAb 12F3 Fab | | Descriptor: | IgE 12F3 Fab heavy chain, IgE 12F3 Fab light chain, Major allergen Can f 1 | | Authors: | Khatri, K, Ball, A, Smith, S.A, Champan, M.D, Pomes, A, Chruszcz, M. | | Deposit date: | 2025-03-11 | | Release date: | 2025-08-27 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Human IgE monoclonal antibodies define two unusual epitopes trapping dog allergen Can f 1 in different conformations.
Protein Sci., 34, 2025
|
|
9Q9L
 
 | | HSV-1 prefusion glycoprotein B | | Descriptor: | Glycoprotein B | | Authors: | Vollmer, B, Mulvaney, T, Ebel, H, Nentwig, J, Gruenewald, K. | | Deposit date: | 2025-02-26 | | Release date: | 2025-09-03 | | Last modified: | 2025-09-17 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | A nanobody specific to prefusion glycoprotein B neutralizes HSV-1 and HSV-2.
Nature, 2025
|
|
9OH6
 
 | | H117A/M121H Azurin with Cu(II), pH 7.7 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Azurin, COPPER (II) ION | | Authors: | Van Stappen, C, Lu, Y. | | Deposit date: | 2025-05-02 | | Release date: | 2025-07-23 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Beyond Blue: Systematic Modulation of Electronic Structure and Redox Properties of Type 1 Copper in Azurin.
J.Am.Chem.Soc., 147, 2025
|
|
9SOP
 
 | | Tissue inhibitor of metalloproteinase-1 (TIMP-1) | | Descriptor: | 3,6,9,12,15-PENTAOXAHEPTADECANE, CITRATE ANION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Shemy, A, Voet, A. | | Deposit date: | 2025-09-15 | | Release date: | 2025-09-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Tissue inhibitor of metalloproteinase-1 (TIMP-1)
To Be Published
|
|
8D1R
 
 | | Crystal structure of acetyltransferase Eis from Mycobacterium tuberculosis in complex with inhibitor SGT520 | | Descriptor: | 1-{3-[(4-chlorophenyl)methoxy]phenyl}methanamine, GLYCEROL, N-acetyltransferase Eis | | Authors: | Pang, A.H, Punetha, A, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2022-05-27 | | Release date: | 2022-09-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Discovery of substituted benzyloxy-benzylamine inhibitors of acetyltransferase Eis and their anti-mycobacterial activity.
Eur.J.Med.Chem., 242, 2022
|
|
