7PM7
 
 | | Cryo-EM structure of the actomyosin-V complex in the strong-ADP state (central 1er, class 2) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Pospich, S, Sweeney, H.L, Houdusse, A, Raunser, S. | | Deposit date: | 2021-09-02 | | Release date: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | High-resolution structures of the actomyosin-V complex in three nucleotide states provide insights into the force generation mechanism.
Elife, 10, 2021
|
|
7PLW
 
 | | Cryo-EM structure of the actomyosin-V complex in the rigor state (central 1er, class 2) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Pospich, S, Sweeney, H.L, Houdusse, A, Raunser, S. | | Deposit date: | 2021-09-01 | | Release date: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | High-resolution structures of the actomyosin-V complex in three nucleotide states provide insights into the force generation mechanism.
Elife, 10, 2021
|
|
7PMJ
 
 | | Cryo-EM structure of the actomyosin-V complex in the post-rigor transition state (AppNHp, central 1er, class 6) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Pospich, S, Sweeney, H.L, Houdusse, A, Raunser, S. | | Deposit date: | 2021-09-02 | | Release date: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | High-resolution structures of the actomyosin-V complex in three nucleotide states provide insights into the force generation mechanism.
Elife, 10, 2021
|
|
7PM6
 
 | | Cryo-EM structure of the actomyosin-V complex in the strong-ADP state (central 3er/2er) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Pospich, S, Sweeney, H.L, Houdusse, A, Raunser, S. | | Deposit date: | 2021-09-02 | | Release date: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | High-resolution structures of the actomyosin-V complex in three nucleotide states provide insights into the force generation mechanism.
Elife, 10, 2021
|
|
8WM9
 
 | | Fzd4/DEP complex | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Frizzled-4, Segment polarity protein dishevelled homolog DVL-2 | | Authors: | He, Y, Qian, Y. | | Deposit date: | 2023-10-03 | | Release date: | 2024-09-11 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Structural basis of Frizzled 4 in recognition of Dishevelled 2 unveils mechanism of WNT signaling activation.
Nat Commun, 15, 2024
|
|
7PMA
 
 | | Cryo-EM structure of the actomyosin-V complex in the strong-ADP state (central 1er, class 5) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Pospich, S, Sweeney, H.L, Houdusse, A, Raunser, S. | | Deposit date: | 2021-09-02 | | Release date: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | High-resolution structures of the actomyosin-V complex in three nucleotide states provide insights into the force generation mechanism.
Elife, 10, 2021
|
|
7PM0
 
 | | Cryo-EM structure of the actomyosin-V complex in the rigor state (central 1er, young JASP-stabilized F-actin, class 1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Pospich, S, Sweeney, H.L, Houdusse, A, Raunser, S. | | Deposit date: | 2021-09-01 | | Release date: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | High-resolution structures of the actomyosin-V complex in three nucleotide states provide insights into the force generation mechanism.
Elife, 10, 2021
|
|
7PMC
 
 | | Cryo-EM structure of the actomyosin-V complex in the strong-ADP state (central 1er, class 7) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Pospich, S, Sweeney, H.L, Houdusse, A, Raunser, S. | | Deposit date: | 2021-09-02 | | Release date: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | High-resolution structures of the actomyosin-V complex in three nucleotide states provide insights into the force generation mechanism.
Elife, 10, 2021
|
|
7PJD
 
 | |
8WOG
 
 | | Cryo-EM structure of SUCR1 in complex with succinate and Gi protein | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Liu, A, Ye, R.D. | | Deposit date: | 2023-10-07 | | Release date: | 2024-09-11 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structural insights into ligand recognition and activation of the succinate receptor SUCNR1.
Cell Rep, 43, 2024
|
|
7PLY
 
 | | Cryo-EM structure of the actomyosin-V complex in the rigor state (central 1er, young JASP-stabilized F-actin) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Pospich, S, Sweeney, H.L, Houdusse, A, Raunser, S. | | Deposit date: | 2021-09-01 | | Release date: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | High-resolution structures of the actomyosin-V complex in three nucleotide states provide insights into the force generation mechanism.
Elife, 10, 2021
|
|
7PMB
 
 | | Cryo-EM structure of the actomyosin-V complex in the strong-ADP state (central 1er, class 6) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Pospich, S, Sweeney, H.L, Houdusse, A, Raunser, S. | | Deposit date: | 2021-09-02 | | Release date: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | High-resolution structures of the actomyosin-V complex in three nucleotide states provide insights into the force generation mechanism.
Elife, 10, 2021
|
|
7PLT
 
 | | Cryo-EM structure of the actomyosin-V complex in the rigor state (central 1er) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Pospich, S, Sweeney, H.L, Houdusse, A, Raunser, S. | | Deposit date: | 2021-09-01 | | Release date: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | High-resolution structures of the actomyosin-V complex in three nucleotide states provide insights into the force generation mechanism.
Elife, 10, 2021
|
|
7PM5
 
 | | Cryo-EM structure of the actomyosin-V complex in the strong-ADP state (central 1er) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Pospich, S, Sweeney, H.L, Houdusse, A, Raunser, S. | | Deposit date: | 2021-09-02 | | Release date: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | High-resolution structures of the actomyosin-V complex in three nucleotide states provide insights into the force generation mechanism.
Elife, 10, 2021
|
|
7PMI
 
 | | Cryo-EM structure of the actomyosin-V complex in the post-rigor transition state (AppNHp, central 1er, class 5) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Pospich, S, Sweeney, H.L, Houdusse, A, Raunser, S. | | Deposit date: | 2021-09-02 | | Release date: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | High-resolution structures of the actomyosin-V complex in three nucleotide states provide insights into the force generation mechanism.
Elife, 10, 2021
|
|
8WJX
 
 | |
7PFS
 
 | | Crystal structure of ERAP2 aminopeptidase in complex with phosphinic pseudotripeptide ((1R)-1-Amino-3-phenylpropyl){2-([1,1:3,1-terphenyl]-5-ylmethyl)-3-[((2S)-1-amino-1-oxo-3-phenylpropan-2-yl)-amino]-3-oxopropyl}phosphinic acid | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Giastas, P, Stratikos, E, Mpakali, A. | | Deposit date: | 2021-08-12 | | Release date: | 2022-01-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Inhibitor-Dependent Usage of the S1' Specificity Pocket of ER Aminopeptidase 2.
Acs Med.Chem.Lett., 13, 2022
|
|
7P7P
 
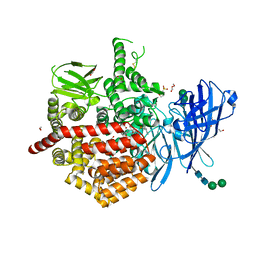 | | Crystal structure of ERAP2 aminopeptidase in complex with phosphinic pseudotripeptide((1R)-1-Amino-3-phenylpropyl){(2S)-3-[((2S)-1-amino-1-oxo-3-phenylpropan-2-yl)amino]-2-{[3-(2-hydroxyphenyl)-isoxazol-5-yl]methyl}-3-oxopropyl}phosphinic acid | | Descriptor: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Giastas, P, Stratikos, E, Mpakali, A. | | Deposit date: | 2021-07-20 | | Release date: | 2022-01-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Inhibitor-Dependent Usage of the S1' Specificity Pocket of ER Aminopeptidase 2.
Acs Med.Chem.Lett., 13, 2022
|
|
3U7X
 
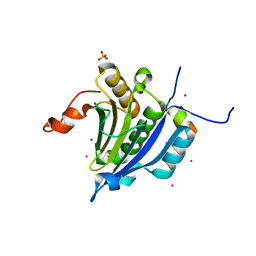 | | Crystal structure of the human eIF4E-4EBP1 peptide complex without cap | | Descriptor: | Eukaryotic translation initiation factor 4E, Eukaryotic translation initiation factor 4E-binding protein 1, SULFATE ION, ... | | Authors: | Siddiqui, N, Tempel, W, Nedyalkova, L, Wernimont, A.K, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Borden, K.L.B, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-10-14 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Insights into the Allosteric Effects of 4EBP1 on the Eukaryotic Translation Initiation Factor eIF4E.
J.Mol.Biol., 415, 2012
|
|
3U3I
 
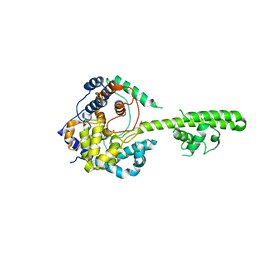 | | A RNA binding protein from Crimean-Congo hemorrhagic fever virus | | Descriptor: | Nucleocapsid protein | | Authors: | Guo, Y, Wang, W.M, Ji, W, Deng, M, Sun, Y.N, Lou, Z.Y, Rao, Z.H. | | Deposit date: | 2011-10-06 | | Release date: | 2012-03-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.304 Å) | | Cite: | Crimean-Congo hemorrhagic fever virus nucleoprotein reveals endonuclease activity in bunyaviruses
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
7PKY
 
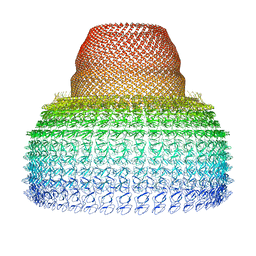 | | Half-vault structure | | Descriptor: | Major vault protein | | Authors: | Guerra, P, Gonzalez-Alamos, M, Llauro, A, Casanas, A, Querol-Audi, J, de Pablo, P, Verdaguer, N. | | Deposit date: | 2021-08-27 | | Release date: | 2022-03-23 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Symmetry disruption commits vault particles to disassembly.
Sci Adv, 8, 2022
|
|
3U8U
 
 | |
7PUB
 
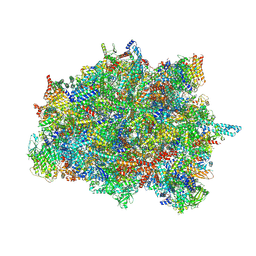 | | Late assembly intermediate of the Trypanosoma brucei mitoribosomal small subunit | | Descriptor: | 30S Ribosomal protein S17, putative, 30S ribosomal protein S8, ... | | Authors: | Lenarcic, T, Leibundgut, M, Saurer, M, Ramrath, D.J.F, Fluegel, T, Boehringer, D, Ban, N. | | Deposit date: | 2021-09-29 | | Release date: | 2022-05-04 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Mitoribosomal small subunit maturation involves formation of initiation-like complexes.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
3TPQ
 
 | |
7PDV
 
 | |
