4ARC
 
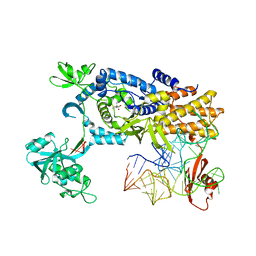 | | Ternary complex of E. coli leucyl-tRNA synthetase, tRNA(leu) and leucine in the editing conformation | | Descriptor: | LEUCINE, LEUCINE--TRNA LIGASE, MAGNESIUM ION, ... | | Authors: | Palencia, A, Crepin, T, Vu, M.T, Lincecum Jr, T.L, Martinis, S.A, Cusack, S. | | Deposit date: | 2012-04-23 | | Release date: | 2012-06-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Dynamics of the Aminoacylation and Proofreading Functional Cycle of Bacterial Leucyl-tRNA Synthetase
Nat.Struct.Mol.Biol., 19, 2012
|
|
2HMK
 
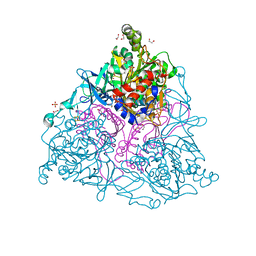 | | Crystal Structure of Naphthalene 1,2-Dioxygenase Bound to Phenanthrene | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Ferraro, D.J, Okerlund, A.L, Mowers, J.C, Ramaswamy, S. | | Deposit date: | 2006-07-11 | | Release date: | 2006-10-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for regioselectivity and stereoselectivity of product formation by naphthalene 1,2-dioxygenase.
J.Bacteriol., 188, 2006
|
|
1TK2
 
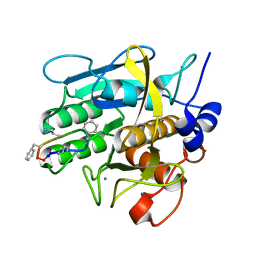 | | Crystal Structure of the Complex formed between Alkaline Proteinase Savinase and Gramicidin S at 1.5A Resolution | | Descriptor: | CALCIUM ION, GRAMICIDIN S, SUBTILISIN SAVINASE | | Authors: | Bhatt, V.S, Kaur, P, Klupsch, S, Betzel, C, Brenner, S, Singh, T.P. | | Deposit date: | 2004-06-08 | | Release date: | 2004-06-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Crystal Structure of the Complex Formed between Alkaline Proteinase Savinase and Gramicidin S at 1.5A Resolution.
To be Published
|
|
1ORP
 
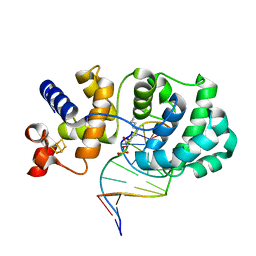 | |
1T0I
 
 | | YLR011wp, a Saccharomyces cerevisiae NA(D)PH-dependent FMN reductase | | Descriptor: | CALCIUM ION, FLAVIN MONONUCLEOTIDE, YLR011wp | | Authors: | Liger, D, Graille, M, Zhou, C.-Z, Leulliot, N, Quevillon-Cheruel, S, Blondeau, K, Janin, J, van Tilbeurgh, H. | | Deposit date: | 2004-04-09 | | Release date: | 2004-06-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure and Functional Characterization of Yeast YLR011wp, an Enzyme with NAD(P)H-FMN and Ferric Iron Reductase Activities
J.Biol.Chem., 279, 2004
|
|
2HMN
 
 | | Crystal Structure of the Naphthalene 1,2-Dioxygenase F352V Mutant Bound to Anthracene. | | Descriptor: | 1,2-ETHANEDIOL, ANTHRACENE, FE (III) ION, ... | | Authors: | Ferraro, D.J, Okerlund, A.L, Mowers, J.C, Ramaswamy, S. | | Deposit date: | 2006-07-11 | | Release date: | 2006-10-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for regioselectivity and stereoselectivity of product formation by naphthalene 1,2-dioxygenase.
J.Bacteriol., 188, 2006
|
|
3LGN
 
 | | Crystal structure of IsdI in complex with heme | | Descriptor: | Heme-degrading monooxygenase isdI, MAGNESIUM ION, OXYGEN MOLECULE, ... | | Authors: | Ukpabi, G.N, Murphy, M.E.P. | | Deposit date: | 2010-01-20 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The IsdG-family of haem oxygenases degrades haem to a novel chromophore
Mol.Microbiol., 75, 2010
|
|
2AFO
 
 | | Crystal structure of human glutaminyl cyclase at pH 8.0 | | Descriptor: | Glutaminyl-peptide cyclotransferase, SULFATE ION, ZINC ION | | Authors: | Huang, K.F, Liu, Y.L, Cheng, W.J, Ko, T.P, Wang, A.H.J. | | Deposit date: | 2005-07-26 | | Release date: | 2005-08-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structures of human glutaminyl cyclase, an enzyme responsible for protein N-terminal pyroglutamate formation
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2AFW
 
 | | Crystal structure of human glutaminyl cyclase in complex with N-acetylhistamine | | Descriptor: | Glutaminyl-peptide cyclotransferase, N-[2-(1H-IMIDAZOL-4-YL)ETHYL]ACETAMIDE, SULFATE ION, ... | | Authors: | Huang, K.F, Liu, Y.L, Cheng, W.J, Ko, T.P, Wang, A.H.J. | | Deposit date: | 2005-07-26 | | Release date: | 2005-08-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Crystal structures of human glutaminyl cyclase, an enzyme responsible for protein N-terminal pyroglutamate formation
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1SBT
 
 | | ATOMIC COORDINATES FOR SUBTILISIN BPN (OR NOVO) | | Descriptor: | SUBTILISIN BPN' | | Authors: | Alden, R.A, Birktoft, J.J, Kraut, J, Robertus, J.D, Wright, C.S. | | Deposit date: | 1972-08-11 | | Release date: | 1977-01-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Atomic coordinates for subtilisin BPN' (or Novo).
Biochem.Biophys.Res.Commun., 45, 1971
|
|
1SBI
 
 | | SUBTILISIN BPN' 8397 (E.C. 3.4.21.14) MUTANT (M50F, N76D, G169A, Q206C, N218S) | | Descriptor: | CALCIUM ION, SUBTILISIN 8397 | | Authors: | Kidd, R.D, Yennawar, H.P, Farber, G.K. | | Deposit date: | 1995-09-01 | | Release date: | 1995-12-07 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A weak calcium binding site in subtilisin BPN' has a dramatic effect on protein stability.
J.Am.Chem.Soc., 118, 1996
|
|
1SUB
 
 | | CALCIUM-INDEPENDENT SUBTILISIN BY DESIGN | | Descriptor: | ACETONE, CALCIUM ION, POTASSIUM ION, ... | | Authors: | Gallagher, T, Bryan, P, Gilliland, G.L. | | Deposit date: | 1992-06-10 | | Release date: | 1994-01-31 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Calcium-independent subtilisin by design.
Proteins, 16, 1993
|
|
2AB6
 
 | |
2A1N
 
 | |
2KA5
 
 | | NMR Structure of the protein TM1081 | | Descriptor: | Putative anti-sigma factor antagonist TM_1081 | | Authors: | Serrano, P, Geralt, M, Mohanty, B, Pedrini, B, Horst, R, Wuthrich, K, Wilson, I, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2008-10-30 | | Release date: | 2008-11-25 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Comparison of NMR and crystal structures highlights conformational isomerism in protein active sites.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
1Q13
 
 | | Crystal structure of rabbit 20alpha hyroxysteroid dehydrogenase in ternary complex with NADP and testosterone | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Prostaglandin-E2 9-reductase, SULFATE ION, ... | | Authors: | Couture, J.-F, Cantin, L, Legrand, P, Luu-The, V, Labrie, F, Breton, R. | | Deposit date: | 2003-07-18 | | Release date: | 2004-11-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Loop relaxation, a mechanism that explains the reduced specificity of rabbit 20alpha-hydroxysteroid dehydrogenase, a member of the aldo-keto reductase superfamily.
J. Mol. Biol., 339, 2004
|
|
1W5L
 
 | | An anti-parallel to parallel switch. | | Descriptor: | GENERAL CONTROL PROTEIN GCN4 | | Authors: | Yadav, M.K, Leman, L.J, Stout, C.D, Ghadiri, M.R. | | Deposit date: | 2004-08-07 | | Release date: | 2004-09-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Coiled Coils at the Edge of Configurational Heterogeneity. Structural Analyses of Parallel and Antiparallel Homotetrameric Coiled Coils Reveal Configurational Sensitivity to a Single Solvent-Exposed Amino Acid Substitution.
Biochemistry, 45, 2006
|
|
1VRN
 
 | | PHOTOSYNTHETIC REACTION CENTER BLASTOCHLORIS VIRIDIS (ATCC) | | Descriptor: | 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, ... | | Authors: | Baxter, R.H.G, Seagle, B.-L, Norris, J.R. | | Deposit date: | 2005-02-23 | | Release date: | 2005-03-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Cryogenic structure of the photosynthetic reaction center of Blastochloris viridis in the light and dark.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2FYM
 
 | |
1P28
 
 | | The crystal structure of a pheromone binding protein from the cockroach Leucophaea maderae in complex with a component of the pheromonal blend: 3-hydroxy-butan-2-one. | | Descriptor: | R,3-HYDROXYBUTAN-2-ONE, S,3-HYDROXYBUTAN-2-ONE, pheromone binding protein | | Authors: | Lartigue, A, Gruez, A, Spinelli, S, Riviere, S, Brossut, R, Tegoni, M, Cambillau, C. | | Deposit date: | 2003-04-15 | | Release date: | 2003-08-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | THE CRYSTAL STRUCTURE OF A COCKROACH PHEROMONE-BINDING PROTEIN SUGGESTS A NEW LIGAND BINDING AND RELEASE MECHANISM
J.Biol.Chem., 278, 2003
|
|
1ORN
 
 | |
2AF0
 
 | | Structure of the Regulator of G-Protein Signaling Domain of RGS2 | | Descriptor: | Regulator of G-protein signaling 2 | | Authors: | Papagrigoriou, E, Johannson, C, Phillips, C, Smee, C, Elkins, J.M, Weigelt, J, Arrowsmith, C, Edwards, A, Sundstrom, M, Von Delft, F, Doyle, D.A, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-07-25 | | Release date: | 2005-08-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural diversity in the RGS domain and its interaction with heterotrimeric G protein alpha-subunits.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2AFZ
 
 | | Crystal structure of human glutaminyl cyclase in complex with 1-vinylimidazole | | Descriptor: | 1-VINYLIMIDAZOLE, Glutaminyl-peptide cyclotransferase, SULFATE ION, ... | | Authors: | Huang, K.F, Liu, Y.L, Cheng, W.J, Ko, T.P, Wang, A.H.J. | | Deposit date: | 2005-07-26 | | Release date: | 2005-08-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal structures of human glutaminyl cyclase, an enzyme responsible for protein N-terminal pyroglutamate formation
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2AFS
 
 | | Crystal structure of the genetic mutant R54W of human glutaminyl cyclase | | Descriptor: | Glutaminyl-peptide cyclotransferase, SULFATE ION, ZINC ION | | Authors: | Huang, K.F, Liu, Y.L, Cheng, W.J, Ko, T.P, Wang, A.H.J. | | Deposit date: | 2005-07-26 | | Release date: | 2005-08-23 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal structures of human glutaminyl cyclase, an enzyme responsible for protein N-terminal pyroglutamate formation
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1ORG
 
 | | The crystal structure of a pheromone binding protein from the cockroach Leucophaea maderae reveals a new mechanism of pheromone binding | | Descriptor: | GLYCEROL, pheromone binding protein | | Authors: | Lartigue, A, Gruez, A, Spinelli, S, Riviere, S, Brossut, R, Tegoni, M, Cambillau, C. | | Deposit date: | 2003-03-13 | | Release date: | 2003-08-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | THE CRYSTAL STRUCTURE OF A COCKROACH PHEROMONE-BINDING PROTEIN SUGGESTS A NEW LIGAND BINDING AND RELEASE MECHANISM
J.Biol.Chem., 278, 2003
|
|
