5R9B
 
 | |
5R9S
 
 | |
6Y8C
 
 | |
6W21
 
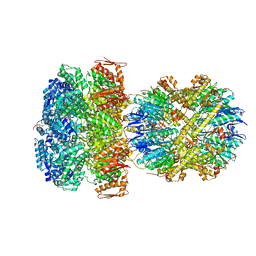 | | ClpAP Engaged2 State bound to RepA-GFP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ... | | Authors: | Lopez, K.L, Rizo, A.N, Tse, E, Lin, J, Scull, N.W, Thwin, A.C, Lucius, A.L, Shorter, J, Southworth, D.R. | | Deposit date: | 2020-03-04 | | Release date: | 2020-05-13 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Conformational plasticity of the ClpAP AAA+ protease couples protein unfolding and proteolysis.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6Y89
 
 | | Crystal structure of the cAMP-dependent protein kinase A cocrystallized with Methyl 5-isoquinolinecarboxylate and PKI (5-24) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, METHANOL, ... | | Authors: | Oebbeke, M, Gerber, H.-D, Heine, A, Klebe, G. | | Deposit date: | 2020-03-04 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Fragment based drug design - Small chemical changes of fragments effecting big changes in binding
To Be Published
|
|
6W22
 
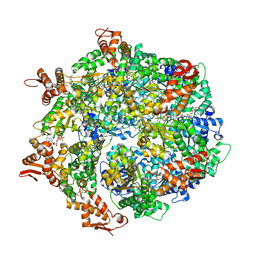 | | ClpA Engaged1 State bound to RepA-GFP (ClpA Focused Refinement) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ... | | Authors: | Lopez, K.L, Rizo, A.N, Tse, E, Lin, J, Scull, N.W, Thwin, A.C, Lucius, A.L, Shorter, J, Southworth, D.R. | | Deposit date: | 2020-03-04 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Conformational plasticity of the ClpAP AAA+ protease couples protein unfolding and proteolysis.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6W25
 
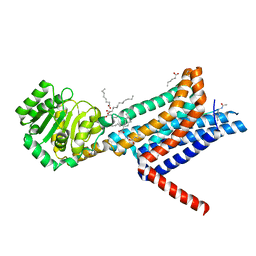 | | Crystal structure of the Melanocortin-4 Receptor (MC4R) in complex with SHU9119 | | Descriptor: | CALCIUM ION, Melanocortin receptor 4,GlgA glycogen synthase,Melanocortin receptor 4, OLEIC ACID, ... | | Authors: | Yu, J, Gimenez, L.E, Hernandez, C.C, Wu, Y, Wein, A.H, Han, G.W, McClary, K, Mittal, S.R, Burdsall, K, Stauch, B, Wu, L, Stevens, S.N, Peisley, A, Williams, S.Y, Chen, V, Millhauser, G.L, Zhao, S, Cone, R.D, Stevens, R.C. | | Deposit date: | 2020-03-04 | | Release date: | 2020-04-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Determination of the melanocortin-4 receptor structure identifies Ca2+as a cofactor for ligand binding.
Science, 368, 2020
|
|
6W23
 
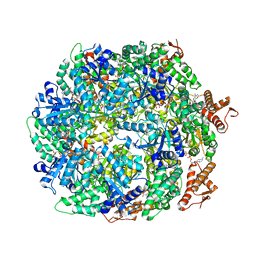 | | ClpA Disengaged State bound to RepA-GFP (Focused Classification) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ... | | Authors: | Lopez, K.L, Rizo, A.N, Tse, E, Lin, J, Scull, N.W, Thwin, A.C, Lucius, A.L, Shorter, J, Southworth, D.R. | | Deposit date: | 2020-03-04 | | Release date: | 2020-05-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Conformational plasticity of the ClpAP AAA+ protease couples protein unfolding and proteolysis.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6W1Z
 
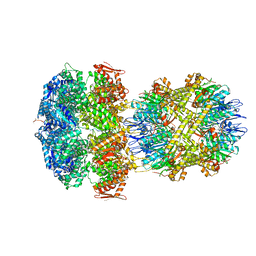 | | ClpAP Engaged1 State bound to RepA-GFP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ... | | Authors: | Lopez, K.L, Rizo, A.N, Tse, E, Lin, J, Scull, N.W, Thwin, A.C, Lucius, A.L, Shorter, J, Southworth, D.R. | | Deposit date: | 2020-03-04 | | Release date: | 2020-05-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Conformational plasticity of the ClpAP AAA+ protease couples protein unfolding and proteolysis.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6W24
 
 | | ClpA Engaged2 State bound to RepA-GFP (Focused Classification) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ... | | Authors: | Lopez, K.L, Rizo, A.N, Tse, E, Lin, J, Scull, N.W, Thwin, A.C, Lucius, A.L, Shorter, J, Southworth, D.R. | | Deposit date: | 2020-03-04 | | Release date: | 2020-05-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Conformational plasticity of the ClpAP AAA+ protease couples protein unfolding and proteolysis.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6W20
 
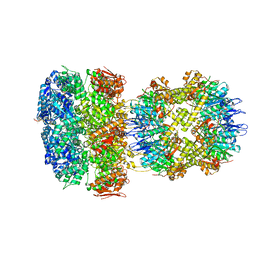 | | ClpAP Disengaged State bound to RepA-GFP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ... | | Authors: | Lopez, K.L, Rizo, A.N, Tse, E, Lin, J, Scull, N.W, Thwin, A.C, Lucius, A.L, Shorter, J, Southworth, D.R. | | Deposit date: | 2020-03-04 | | Release date: | 2020-05-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Conformational plasticity of the ClpAP AAA+ protease couples protein unfolding and proteolysis.
Nat.Struct.Mol.Biol., 27, 2020
|
|
5R93
 
 | |
5R9K
 
 | |
5RA2
 
 | |
5R9L
 
 | |
5RA3
 
 | |
5R94
 
 | |
5R9H
 
 | |
5R8V
 
 | |
5R9Y
 
 | |
5R9A
 
 | |
5R9Q
 
 | |
5RA5
 
 | |
5R91
 
 | |
5R9M
 
 | |
