5SOD
 
 | | PanDDA analysis group deposition -- Crystal Structure of Pseudomonas Aeruginosa FabF-C164Q mutant protein in complex with Z1730522163 | | Descriptor: | 1-[(2R)-8-fluoro-2-methyl-2,3-dihydro-4H-1,4-benzoxazin-4-yl]ethan-1-one, 3-oxoacyl-[acyl-carrier-protein] synthase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Brenk, R, Georgiou, C. | | Deposit date: | 2022-05-30 | | Release date: | 2023-12-20 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Towards new antibiotics: P. aeruginosa FabF ligands discovered by crystallographic fragment screening followed by hit expansion.
Eur.J.Med.Chem., 291, 2025
|
|
5SNI
 
 | |
5SNW
 
 | |
5SN6
 
 | |
5SNN
 
 | |
5SO5
 
 | |
5SOA
 
 | |
5SNK
 
 | |
5SNX
 
 | |
5SOB
 
 | |
5SNE
 
 | |
5SO1
 
 | |
5SOH
 
 | |
2WEW
 
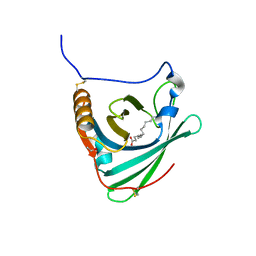 | | Crystal structure of human apoM in complex with myristic acid | | Descriptor: | 1,2-ETHANEDIOL, APOLIPOPROTEIN M, MYRISTIC ACID | | Authors: | Sevvana, M, Ahnstrom, J, Egerer-Sieber, C, Dahlback, B, Muller, Y.A. | | Deposit date: | 2009-04-02 | | Release date: | 2009-09-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Serendipitous Fatty Acid Binding Reveals the Structural Determinants for Ligand Recognition in Apolipoprotein M.
J.Mol.Biol., 393, 2009
|
|
2G0K
 
 | |
4FEN
 
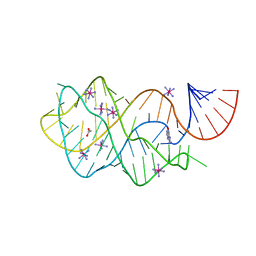 | | Crystal structure of the A24U/U25A/A46G mutant xpt-pbuX guanine riboswitch aptamer domain in complex with hypoxanthine | | Descriptor: | A24U/U25A/A46G mutant of the B. subtilis xpt-pbuX guanine riboswitch aptamer domain, ACETATE ION, COBALT HEXAMMINE(III), ... | | Authors: | Stoddard, C.D, Trausch, J.J, Widmann, J, Marcano, J, Knight, R, Batey, R.T. | | Deposit date: | 2012-05-30 | | Release date: | 2013-02-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Nucleotides Adjacent to the Ligand-Binding Pocket are Linked to Activity Tuning in the Purine Riboswitch.
J.Mol.Biol., 425, 2013
|
|
1Z8O
 
 | | Ferrous dioxygen complex of the wild-type cytochrome P450eryF | | Descriptor: | 6-DEOXYERYTHRONOLIDE B, 6-deoxyerythronolide B hydroxylase, OXYGEN MOLECULE, ... | | Authors: | Nagano, S, Cupp-Vickery, J.R, Poulos, T.L. | | Deposit date: | 2005-03-31 | | Release date: | 2005-04-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of the ferrous dioxygen complex of wild-type cytochrome P450eryF and its mutants, A245S and A245T: investigation of the proton transfer system in P450eryF.
J.Biol.Chem., 280, 2005
|
|
2WFI
 
 | |
2WFJ
 
 | | Atomic resolution crystal structure of the PPIase domain of human cyclophilin G in complex with cyclosporin A. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, CYCLOSPORIN A, ... | | Authors: | Stegmann, C.M, Sheldrick, G.M, Wahl, M.C. | | Deposit date: | 2009-04-06 | | Release date: | 2009-06-16 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (0.75 Å) | | Cite: | The Thermodynamic Influence of Trapped Water Molecules on a Protein-Ligand Interaction.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
6DHH
 
 | | RT XFEL structure of Photosystem II 400 microseconds after the second illumination at 2.2 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kern, J, Chatterjee, R, Young, I.D, Fuller, F.D, Lassalle, L, Ibrahim, M, Gul, S, Fransson, T, Brewster, A.S, Alonso-Mori, R, Hussein, R, Zhang, M, Douthit, L, de Lichtenberg, C, Cheah, M.H, Shevela, D, Wersig, J, Seufert, I, Sokaras, D, Pastor, E, Weninger, C, Kroll, T, Sierra, R.G, Aller, P, Butryn, A, Orville, A.M, Liang, M, Batyuk, A, Koglin, J.E, Carbajo, S, Boutet, S, Moriarty, N.W, Holton, J.M, Dobbek, H, Adams, P.D, Bergmann, U, Sauter, N.K, Zouni, A, Messinger, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2018-05-20 | | Release date: | 2018-11-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of the intermediates of Kok's photosynthetic water oxidation clock.
Nature, 563, 2018
|
|
1WGM
 
 | | Solution structure of the U-box in human ubiquitin conjugation factor E4A | | Descriptor: | Ubiquitin conjugation factor E4A | | Authors: | Saito, K, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the U-box in human ubiquitin conjugation factor E4A
To be Published
|
|
4F6Z
 
 | | Mutagenesis of zinc ligand residue Cys221 reveals plasticity in the IMP-1 metallo-b-lactamase active site | | Descriptor: | Beta-lactamase, CITRATE ANION, ZINC ION | | Authors: | Horton, L.B, Shanker, S, Sankaran, B, Mikulski, R, Brown, N.G, Phillips, K, Lykissa, E, Prasad, B.V.V, Palzkill, T.G. | | Deposit date: | 2012-05-15 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutagenesis of zinc ligand residue Cys221 reveals plasticity in the IMP-1 metallo-beta-lactamase active site
Antimicrob.Agents Chemother., 56, 2012
|
|
4FEP
 
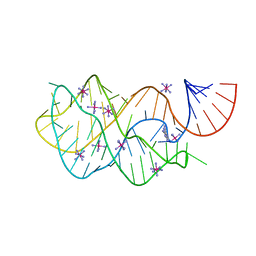 | | Crystal structure of the A24U/U25A/A46G/C74U mutant xpt-pbuX guanine riboswitch aptamer domain in complex with 2,6-diaminopurine | | Descriptor: | 9H-PURINE-2,6-DIAMINE, A24U/U25A/A46G/C74U mutant of the B. subtilis xpt-pbuX guanine riboswitch aptamer domain, COBALT HEXAMMINE(III) | | Authors: | Stoddard, C.D, Trausch, J.J, Widmann, J, Marcano, J, Knight, R, Batey, R.T. | | Deposit date: | 2012-05-30 | | Release date: | 2013-02-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Nucleotides Adjacent to the Ligand-Binding Pocket are Linked to Activity Tuning in the Purine Riboswitch.
J.Mol.Biol., 425, 2013
|
|
1WZ0
 
 | | Solution Structure of Human SUMO-2 (SMT3B), a Ubiquitin-like Protein | | Descriptor: | Ubiquitin-like protein SMT3B | | Authors: | Zhao, C, Saito, K, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-02-21 | | Release date: | 2005-08-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Human SUMO-2 (SMT3B), a Ubiquitin-like Protein
To be Published
|
|
2Q89
 
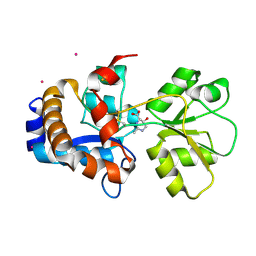 | | Crystal structure of EhuB in complex with hydroxyectoine | | Descriptor: | (4S,5S)-5-HYDROXY-2-METHYL-1,4,5,6-TETRAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, CADMIUM ION, Putative ABC transporter amino acid-binding protein | | Authors: | Hanekop, N, Hoeing, M, Sohn-Bosser, L, Jebbar, M, Schmitt, L, Bremer, E. | | Deposit date: | 2007-06-09 | | Release date: | 2008-01-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the ligand-binding protein EhuB from Sinorhizobium meliloti reveals substrate recognition of the compatible solutes ectoine and hydroxyectoine.
J.Mol.Biol., 374, 2007
|
|
