7DCJ
 
 | |
1BA1
 
 | |
7DCI
 
 | |
7DCU
 
 | | Crystal structure of HSF2 DNA-binding domain in complex with 3-site HSE DNA (21 bp) | | Descriptor: | DNA (5'-D(*AP*CP*CP*GP*CP*GP*AP*AP*TP*AP*TP*TP*CP*TP*AP*GP*AP*AP*CP*GP*C)-3'), DNA (5'-D(*TP*GP*CP*GP*TP*TP*CP*TP*AP*GP*AP*AP*TP*AP*TP*TP*CP*GP*CP*GP*G)-3'), Heat shock factor protein 2, ... | | Authors: | Feng, N, Liu, W. | | Deposit date: | 2020-10-27 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structures of heat shock factor trimers bound to DNA.
Iscience, 24, 2021
|
|
1BGP
 
 | | CRYSTAL STRUCTURE OF BARLEY GRAIN PEROXIDASE 1 | | Descriptor: | BARLEY GRAIN PEROXIDASE, CALCIUM ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Henriksen, A. | | Deposit date: | 1997-07-24 | | Release date: | 1997-11-12 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of barley grain peroxidase refined at 1.9-A resolution. A plant peroxidase reversibly inactivated at neutral pH.
J.Biol.Chem., 273, 1998
|
|
7DCS
 
 | | Crystal structure of HSF1 DNA-binding domain in complex with 3-site HSE DNA (23 bp) | | Descriptor: | DNA (5'-D(*AP*TP*CP*CP*GP*CP*GP*AP*AP*TP*AP*TP*TP*CP*TP*AP*GP*AP*AP*CP*GP*CP*C)-3'), DNA (5'-D(*TP*GP*GP*CP*GP*TP*TP*CP*TP*AP*GP*AP*AP*TP*AP*TP*TP*CP*GP*CP*GP*GP*A)-3'), Heat shock factor protein 1, ... | | Authors: | Feng, N, Liu, W. | | Deposit date: | 2020-10-27 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of heat shock factor trimers bound to DNA.
Iscience, 24, 2021
|
|
7DPH
 
 | | H-Ras Q61H in complex with GppNHp (state 1) after structural transition by humidity control | | Descriptor: | GTPase HRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Taniguchi, H, Matsumoto, S, Kawamura, T, Kumasaka, T, Kataoka, T. | | Deposit date: | 2020-12-19 | | Release date: | 2021-07-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Oncogenic mutations Q61L and Q61H confer active form-like structural features to the inactive state (state 1) conformation of H-Ras protein.
Biochem.Biophys.Res.Commun., 565, 2021
|
|
3VE2
 
 | |
3VFB
 
 | | Crystal Structure of HIV-1 Protease Mutant N88D with novel P1'-Ligands GRL-02031 | | Descriptor: | (3aS,5R,6aR)-hexahydro-2H-cyclopenta[b]furan-5-yl [(1S,2R)-1-benzyl-2-hydroxy-3-([(4-methoxyphenyl)sulfonyl]{[(2R)-5-oxopyrrolidin-2-yl]methyl}amino)propyl]carbamate, CHLORIDE ION, GLYCEROL, ... | | Authors: | Yu, X.X, Wang, Y.F, Chang, Y.C.E, Weber, I.T. | | Deposit date: | 2012-01-09 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Potent antiviral HIV-1 protease inhibitor GRL-02031 adapts to the structures of drug resistant mutants with its P1'-pyrrolidinone ring.
J.Med.Chem., 55, 2012
|
|
3V7Z
 
 | | Carboxypeptidase T with GEMSA | | Descriptor: | (2-GUANIDINOETHYLMERCAPTO)SUCCINIC ACID, CALCIUM ION, Carboxypeptidase T, ... | | Authors: | Kuznetsov, S.A, Timofeev, V.I, Akparov, V.K, Kuranova, I.P. | | Deposit date: | 2011-12-22 | | Release date: | 2012-12-26 | | Last modified: | 2015-09-30 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structural insights into the broad substrate specificity of carboxypeptidase T from Thermoactinomyces vulgaris.
Febs J., 282, 2015
|
|
1BA0
 
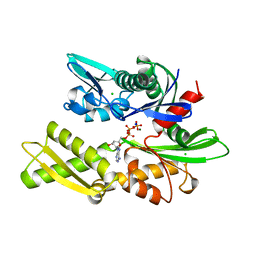 | | HEAT-SHOCK COGNATE 70KD PROTEIN 44KD ATPASE N-TERMINAL 1NGE 3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, HEAT-SHOCK COGNATE 70KD PROTEIN, ... | | Authors: | Wilbanks, S.M, Mckay, D.B. | | Deposit date: | 1998-04-21 | | Release date: | 1998-07-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural replacement of active site monovalent cations by the epsilon-amino group of lysine in the ATPase fragment of bovine Hsc70.
Biochemistry, 37, 1998
|
|
1B7S
 
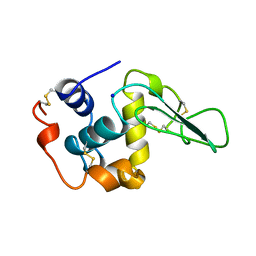 | | VERIFICATION OF SPMP USING MUTANT HUMAN LYSOZYMES | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Takano, K, Ota, M, Ogasahara, K, Yamagata, Y, Nishikawa, K, Yutani, K. | | Deposit date: | 1999-01-25 | | Release date: | 1999-02-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Experimental verification of the 'stability profile of mutant protein' (SPMP) data using mutant human lysozymes.
Protein Eng., 12, 1999
|
|
3VIP
 
 | | Crystal structure of beta-glucosidase from termite Neotermes koshunensis in complex with a new glucopyranosidic product | | Descriptor: | 2-{4-[3-(5H-dibenzo[b,f]azepin-5-yl)propyl]piperazin-1-yl}ethyl beta-D-glucopyranoside, 5H-dibenzo[b,f]azepine, Beta-glucosidase, ... | | Authors: | Jeng, W.Y, Liu, C.I, Wang, A.H.J. | | Deposit date: | 2011-10-03 | | Release date: | 2012-07-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | High-resolution structures of Neotermes koshunensis beta-glucosidase mutants provide insights into the catalytic mechanism and the synthesis of glucoconjugates
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3VW7
 
 | | Crystal structure of human protease-activated receptor 1 (PAR1) bound with antagonist vorapaxar at 2.2 angstrom | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, Proteinase-activated receptor 1, ... | | Authors: | Zhang, C, Srinivasan, Y, Arlow, D.H, Fung, J.J, Palmer, D, Zheng, Y, Green, H.F, Pandey, A, Dror, R.O, Shaw, D.E, Weis, W.I, Coughlin, S.R, Kobilka, B.K. | | Deposit date: | 2012-08-07 | | Release date: | 2012-12-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | High-resolution crystal structure of human protease-activated receptor 1
Nature, 492, 2012
|
|
5XNQ
 
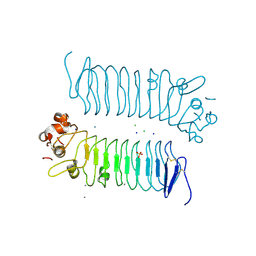 | | Crystal structures of human SALM5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Liu, H, Lin, Z, Xu, F. | | Deposit date: | 2017-05-24 | | Release date: | 2018-01-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Structural basis of SALM5-induced PTP delta dimerization for synaptic differentiation
Nat Commun, 9, 2018
|
|
5XAR
 
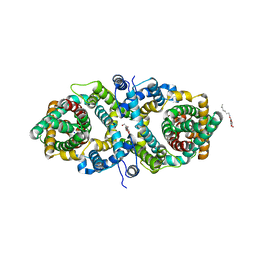 | | Structural insights into the elevator-like mechanism of the sodium/citrate symporter CitS | | Descriptor: | Citrate-sodium symporter, SODIUM ION, octyl beta-D-glucopyranoside | | Authors: | Jin, M.S, Kim, J.W, Kim, S, Kim, S, Lee, H, Lee, J.-O. | | Deposit date: | 2017-03-14 | | Release date: | 2017-06-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.62 Å) | | Cite: | Structural insights into the elevator-like mechanism of the sodium/citrate symporter CitS
Sci Rep, 7, 2017
|
|
3VWP
 
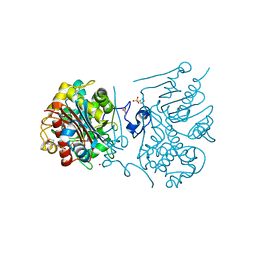 | | Crystal structure of 6-aminohexanoate-dimer hydrolase S112A/G181D/R187S/H266N/D370Y mutant complexd with 6-aminohexanoate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-AMINOHEXANOIC ACID, 6-aminohexanoate-dimer hydrolase, ... | | Authors: | Kawashima, Y, Shibata, N, Negoro, S, Higuchi, Y. | | Deposit date: | 2012-08-30 | | Release date: | 2013-10-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural, kinetic and theoretical analyses of hydrolase mutants altering in the directionality and equilibrium point of reversible amide-synthetic/hydrolytic reaction
to be published
|
|
7DLN
 
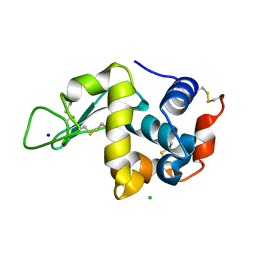 | |
7DLH
 
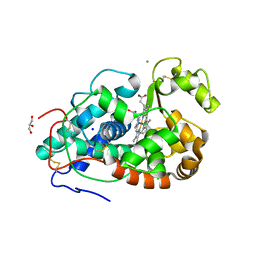 | |
5XHA
 
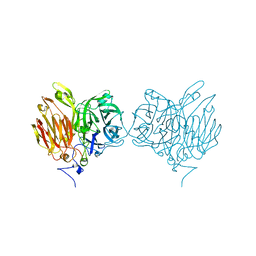 | |
3W1V
 
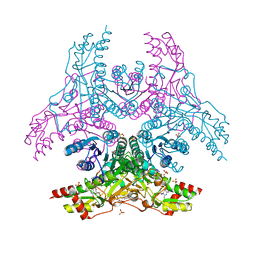 | | Crystal Structure of Capsular Polysaccharide Synthesizing Enzyme CapE from Staphylococcus aureus in complex with inihibitor | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Capsular polysaccharide synthesis enzyme Cap8E, SODIUM ION, ... | | Authors: | Miyafusa, T, Caaveiro, J.M, Tanaka, Y, Tsumoto, K. | | Deposit date: | 2012-11-21 | | Release date: | 2013-06-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the capsular polysaccharide synthesizing protein CapE of Staphylococcus aureus.
Biosci.Rep., 33, 2013
|
|
5X6S
 
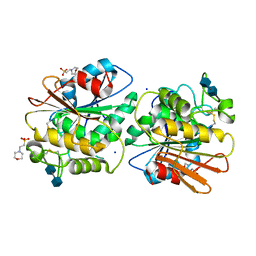 | | Acetyl xylan esterase from Aspergillus awamori | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylxylan esterase A, ... | | Authors: | Komiya, D, Koseki, T, Fushinobu, S. | | Deposit date: | 2017-02-23 | | Release date: | 2017-08-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure and Substrate Specificity Modification of Acetyl Xylan Esterase from Aspergillus luchuensis
Appl. Environ. Microbiol., 83, 2017
|
|
5XAS
 
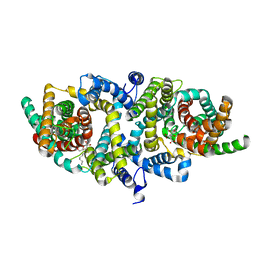 | | Structural insights into the elevator-like mechanism of the sodium/citrate symporter CitS | | Descriptor: | CITRATE ANION, Citrate-sodium symporter, SODIUM ION, ... | | Authors: | Jin, M.S, Kim, J.W, Kim, S, Kim, S, Lee, H, Lee, J.-O. | | Deposit date: | 2017-03-14 | | Release date: | 2017-06-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.47 Å) | | Cite: | Structural insights into the elevator-like mechanism of the sodium/citrate symporter CitS
Sci Rep, 7, 2017
|
|
7EHF
 
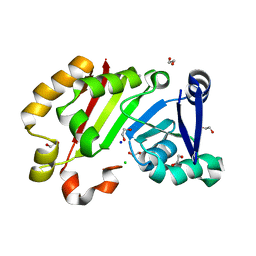 | | Crystal structure of the aminoglycoside resistance methyltransferase NpmB1 | | Descriptor: | 1,2-ETHANEDIOL, 16S rRNA methyltransferase, CHLORIDE ION, ... | | Authors: | Kawai, A, Doi, Y. | | Deposit date: | 2021-03-29 | | Release date: | 2021-08-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Functional and Structural Characterization of Acquired 16S rRNA Methyltransferase NpmB1 Conferring Pan-Aminoglycoside Resistance.
Antimicrob.Agents Chemother., 65, 2021
|
|
1C29
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX OF BACTERIAL TRYPTOPHAN SYNTHASE WITH THE TRANSITION STATE ANALOGUE INHIBITOR 4-(2-HYDROXYPHENYLTHIO)-1-BUTENYLPHOSPHONIC ACID | | Descriptor: | 4-(2-HYDROXYPHENYLTHIO)-1-BUTENYLPHOSPHONIC ACID, PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, ... | | Authors: | Sachpatzidis, A, Dealwis, C, Lubetsky, J.B, Liang, P.H, Anderson, K.S, Lolis, E. | | Deposit date: | 1999-07-23 | | Release date: | 2000-01-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic studies of phosphonate-based alpha-reaction transition-state analogues complexed to tryptophan synthase.
Biochemistry, 38, 1999
|
|
