4MIH
 
 | | Pyranose 2-oxidase from Phanerochaete chrysosporium, recombinant H158A mutant | | Descriptor: | 3-deoxy-3-fluoro-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, Pyranose 2-oxidase | | Authors: | Hassan, N, Tan, T.C, Spadiut, O, Pisanelli, I, Fusco, L, Haltrich, D, Peterbauer, C, Divne, C. | | Deposit date: | 2013-08-31 | | Release date: | 2013-12-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of Phanerochaete chrysosporium pyranose 2-oxidase suggest that the N-terminus acts as a propeptide that assists in homotetramer assembly.
FEBS Open Bio, 3, 2013
|
|
4D1Y
 
 | | Crystal structure of a putative protease from Bacteroides thetaiotaomicron. | | Descriptor: | PUTATIVE PROTEASE I, RIBOFLAVIN, ZINC ION | | Authors: | Knaus, T, Uhl, M.K, Monschein, S, Moratti, S, Gruber, K, Macheroux, P. | | Deposit date: | 2014-05-05 | | Release date: | 2014-10-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and Stability of an Unusual Zinc-Binding Protein from Bacteroides Thetaiotaomicron.
Biochim.Biophys.Acta, 1844, 2014
|
|
3NEA
 
 | | Crystal Structure of Peptidyl-tRNA hydrolase from Francisella tularensis | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Peptidyl-tRNA hydrolase | | Authors: | Lam, R, McGrath, T.E, Romanov, V, Gothe, S.A, Peddi, S.R, Razumova, E, Lipman, R.S, Branstrom, A.A, Chirgadze, N.Y. | | Deposit date: | 2010-06-08 | | Release date: | 2011-06-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of Francisella tularensis peptidyl-tRNA hydrolase.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
4D1D
 
 | | STRUCTURE OF MHP1, A NUCLEOBASE-CATION-SYMPORT-1 FAMILY TRANSPORTER with the inhibitor 5-(2-naphthylmethyl)-L-hydantoin. | | Descriptor: | 5-(2-NAPHTHYLMETHYL)-D-HYDANTOIN, 5-(2-NAPHTHYLMETHYL)-L-HYDANTOIN, HYDANTOIN TRANSPORT PROTEIN, ... | | Authors: | Weyand, S, Brueckner, F, Geng, T, Drew, D, Iwata, S, Henderson, P.J.F, Cameron, A.D. | | Deposit date: | 2014-05-01 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Molecular Mechanism of Ligand Recognition by Membrane Transport Protein, Mhp1.
Embo J., 33, 2014
|
|
3NH3
 
 | |
3NHO
 
 | | Complex of 6-hydroxy-L-nicotine oxidase with product bound at active site | | Descriptor: | (1R)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(pentadecanoyloxy)methyl]ethyl (12E)-hexadeca-9,12-dienoate, 1-(6-hydroxypyridin-3-yl)-4-(methylamino)butan-1-one, 6-hydroxy-L-nicotine oxidase, ... | | Authors: | Kachalova, G.S, Bartunik, H.D. | | Deposit date: | 2010-06-14 | | Release date: | 2011-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structure analysis of free and substrate-bound 6-hydroxy-L-nicotine oxidase from Arthrobacter nicotinovorans.
J.Mol.Biol., 396, 2010
|
|
3NK0
 
 | |
6TME
 
 | | Monomeric LRX8 in complex with RALF4. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(3-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Moussu, S, Caroline, C, Santos-Fernandez, G, Wehrle, S, Grossniklaus, U, Santiago, J. | | Deposit date: | 2019-12-04 | | Release date: | 2020-03-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structural basis for recognition of RALF peptides by LRX proteins during pollen tube growth.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4MIF
 
 | | Pyranose 2-oxidase from Phanerochaete chrysosporium, wild type from natural source | | Descriptor: | DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, MAGNESIUM ION, Pyranose 2-oxidase | | Authors: | Hassan, N, Tan, T.C, Spadiut, O, Pisanelli, I, Fusco, L, Haltrich, D, Peterbauer, C, Divne, C. | | Deposit date: | 2013-08-31 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of Phanerochaete chrysosporium pyranose 2-oxidase suggest that the N-terminus acts as a propeptide that assists in homotetramer assembly.
FEBS Open Bio, 3, 2013
|
|
3NI3
 
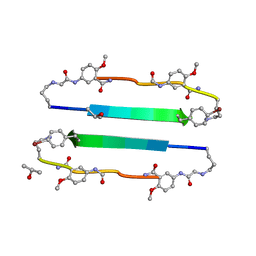 | | 54-Membered ring macrocyclic beta-sheet peptide | | Descriptor: | 54-membered ring macrocyclic beta-sheet peptide, ISOPROPYL ALCOHOL | | Authors: | Sawaya, M.R, Eisenberg, D, Nowick, J.S, Korman, T.P, Khakshoor, O. | | Deposit date: | 2010-06-15 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | X-ray crystallographic structure of an artificial beta-sheet dimer.
J.Am.Chem.Soc., 132, 2010
|
|
6TPV
 
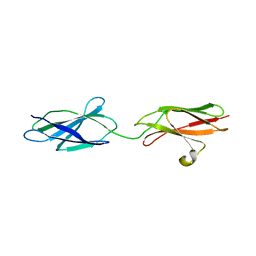 | | Crystal structures of FNIII domain one and two of the human leucocyte common antigen-related protein, LAR | | Descriptor: | IMIDAZOLE, Receptor-type tyrosine-protein phosphatase F | | Authors: | Vilstrup, J.P, Thirup, S.S, Simonsen, A, Birkefeldt, T, Strandbygaard, D. | | Deposit date: | 2019-12-14 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal and solution structures of fragments of the human leucocyte common antigen-related protein.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6TTV
 
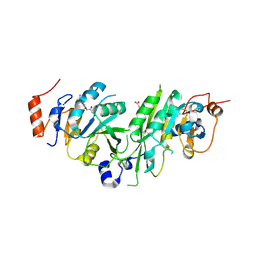 | | Crystal structure of the human METTL3-METTL14 complex bound to Compound 3 (ASI_M3M_138) | | Descriptor: | (2~{S},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolane-2-carboxamide, ACETATE ION, N6-adenosine-methyltransferase catalytic subunit, ... | | Authors: | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | Deposit date: | 2019-12-30 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Small-Molecule Inhibitors of METTL3, the Major Human Epitranscriptomic Writer.
Chemmedchem, 15, 2020
|
|
3N9Y
 
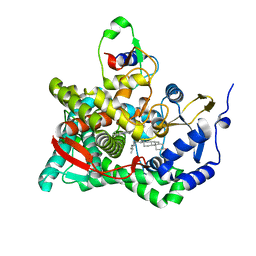 | | Crystal structure of human CYP11A1 in complex with cholesterol | | Descriptor: | Adrenodoxin, CHOLESTEROL, Cholesterol side-chain cleavage enzyme, ... | | Authors: | Strushkevich, N.V, MacKenzie, F, Tempel, W, Botchkarev, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J.U, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-05-31 | | Release date: | 2011-06-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for pregnenolone biosynthesis by the mitochondrial monooxygenase system.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4D1A
 
 | | STRUCTURE OF MHP1, A NUCLEOBASE-CATION-SYMPORT-1 FAMILY TRANSPORTER, IN A CLOSED CONFORMATION WITH INDOLYLMETHYL-HYDANTOIN | | Descriptor: | (5S)-5-(1H-indol-3-ylmethyl)imidazolidine-2,4-dione, HYDANTOIN TRANSPORT PROTEIN, SODIUM ION | | Authors: | Weyand, S, Brueckner, F, Geng, T, Drew, D, Iwata, S, Henderson, P.J.F, Cameron, A.D. | | Deposit date: | 2014-05-01 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Molecular Mechanism of Ligand Recognition by Membrane Transport Protein, Mhp1.
Embo J., 33, 2014
|
|
3N8D
 
 | | Crystal structure of Staphylococcus aureus VRSA-9 D-Ala:D-Ala ligase | | Descriptor: | CHLORIDE ION, D-alanine--D-alanine ligase, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Saul, F.A, Haouz, A, Meziane-Cherif, D. | | Deposit date: | 2010-05-28 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis of vancomycin dependence in VanA-type Staphylococcus aureus VRSA-9.
J.Bacteriol., 192, 2010
|
|
4D1B
 
 | | STRUCTURE OF MHP1, A NUCLEOBASE-CATION-SYMPORT-1 FAMILY TRANSPORTER, IN A CLOSED CONFORMATION WITH BENZYL-HYDANTOIN | | Descriptor: | (5S)-5-benzylimidazolidine-2,4-dione, HYDANTOIN TRANSPORT PROTEIN, SODIUM ION | | Authors: | Brueckner, F, Geng, T, Weyand, S, Drew, D, Iwata, S, Henderson, P.J.F, Cameron, A.D. | | Deposit date: | 2014-05-01 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Molecular Mechanism of Ligand Recognition by Membrane Transport Protein, Mhp1.
Embo J., 33, 2014
|
|
4D6X
 
 | | Crystal structure of the receiver domain of NtrX from Brucella abortus | | Descriptor: | BACTERIAL REGULATORY, FIS FAMILY PROTEIN, IMIDAZOLE | | Authors: | Klinke, S, Fernandez, I, Carrica, M.C, Otero, L.H, Goldbaum, F.A. | | Deposit date: | 2014-11-18 | | Release date: | 2015-07-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Snapshots of Conformational Changes Shed Light Into the Ntrx Receiver Domain Signal Transduction Mechanism
J.Mol.Biol., 427, 2015
|
|
4D3G
 
 | | Structure of PstA | | Descriptor: | PSTA | | Authors: | Campeotto, I, Freemont, P, Grundling, A. | | Deposit date: | 2014-10-22 | | Release date: | 2014-12-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Complex Structure and Biochemical Characterization of the Staphylococcus Aureus Cyclic Di-AMP Binding Protein Psta, the Founding Member of a New Signal Transduction Protein Family
J.Biol.Chem., 290, 2015
|
|
6THL
 
 | | Crystal structure of the complex between RTT106 and BCD1 | | Descriptor: | Box C/D snoRNA protein 1, Rtt106p | | Authors: | Charron, C, Bragantini, B, Manival, X, Charpentier, B. | | Deposit date: | 2019-11-20 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The box C/D snoRNP assembly factor Bcd1 interacts with the histone chaperone Rtt106 and controls its transcription dependent activity.
Nat Commun, 12, 2021
|
|
4CX8
 
 | | Monomeric pseudorabies virus protease pUL26N at 2.5 A resolution | | Descriptor: | PSEUDORABIES VIRUS PROTEASE | | Authors: | Zuehlsdorf, M, Werten, S, Palm, G.J, Hinrichs, W. | | Deposit date: | 2014-04-04 | | Release date: | 2015-05-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Dimerization-Induced Allosteric Changes of the Oxyanion-Hole Loop Activate the Pseudorabies Virus Assemblin Pul26N, a Herpesvirus Serine Protease.
Plos Pathog., 11, 2015
|
|
6OI4
 
 | | RPN13 (19-132)-RPN2 (940-952) pY950-Ub complex | | Descriptor: | 1,2-ETHANEDIOL, 26S proteasome non-ATPase regulatory subunit 1, Proteasomal ubiquitin receptor ADRM1, ... | | Authors: | Hemmis, C.W, Hill, C.P. | | Deposit date: | 2019-04-08 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Phosphorylation of Tyr-950 in the proteasome scaffolding protein RPN2 modulates its interaction with the ubiquitin receptor RPN13.
J.Biol.Chem., 294, 2019
|
|
6DF1
 
 | |
6OKT
 
 | |
6TM6
 
 | | MUC2 CysD1 domain | | Descriptor: | CALCIUM ION, Mucin-2 | | Authors: | Khmelnitsky, L, Fass, D. | | Deposit date: | 2019-12-03 | | Release date: | 2020-02-19 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Assembly Mechanism of Mucin and von Willebrand Factor Polymers.
Cell, 183, 2020
|
|
3N8X
 
 | | Crystal Structure of Cyclooxygenase-1 in Complex with Nimesulide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-NITRO-2-PHENOXYMETHANESULFONANILIDE, ... | | Authors: | Lee, J.Y. | | Deposit date: | 2010-05-28 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Comparison of Cyclooxygenase-1 Crystal Structures: Cross-Talk between Monomers Comprising Cyclooxygenase-1 Homodimers
Biochemistry, 49, 2010
|
|
