1DJF
 
 | | NMR STRUCTURE OF A MODEL HYDROPHILIC AMPHIPATHIC HELICAL BASIC PEPTIDE | | Descriptor: | GLN-ALA-PRO-ALA-TYR-LYS-LYS-ALA-ALA-LYS-LYS-LEU-ALA-GLU-SER | | Authors: | Montserret, R, McLeish, M.J, Bockmann, A, Geourjon, C, Penin, F. | | Deposit date: | 1999-12-03 | | Release date: | 1999-12-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Involvement of electrostatic interactions in the mechanism of peptide folding induced by sodium dodecyl sulfate binding.
Biochemistry, 39, 2000
|
|
1DJ8
 
 | |
1QR0
 
 | | CRYSTAL STRUCTURE OF THE 4'-PHOSPHOPANTETHEINYL TRANSFERASE SFP-COENZYME A COMPLEX | | Descriptor: | 4'-PHOSPHOPANTETHEINYL TRANSFERASE SFP, COENZYME A, MAGNESIUM ION | | Authors: | Reuter, K, Mofid, R.M, Marahiel, A.M, Ficner, R. | | Deposit date: | 1999-06-17 | | Release date: | 1999-12-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the surfactin synthetase-activating enzyme sfp: a prototype of the 4'-phosphopantetheinyl transferase superfamily.
EMBO J., 18, 1999
|
|
1QUP
 
 | | CRYSTAL STRUCTURE OF THE COPPER CHAPERONE FOR SUPEROXIDE DISMUTASE | | Descriptor: | SULFATE ION, SUPEROXIDE DISMUTASE 1 COPPER CHAPERONE | | Authors: | Lamb, A.L, Wernimont, A.K, Pufahl, R.A, O'Halloran, T.V, Rosenzweig, A.C. | | Deposit date: | 1999-07-01 | | Release date: | 1999-12-10 | | Last modified: | 2018-06-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the copper chaperone for superoxide dismutase.
Nat.Struct.Biol., 6, 1999
|
|
1B3K
 
 | | Plasminogen activator inhibitor-1 | | Descriptor: | PLASMINOGEN ACTIVATOR INHIBITOR-1 | | Authors: | Sharp, A.M, Stein, P.E, Pannu, N.S, Read, R.J. | | Deposit date: | 1998-12-11 | | Release date: | 1999-12-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | The active conformation of plasminogen activator inhibitor 1, a target for drugs to control fibrinolysis and cell adhesion.
Structure Fold.Des., 7, 1999
|
|
1C8Z
 
 | |
1CC8
 
 | | CRYSTAL STRUCTURE OF THE ATX1 METALLOCHAPERONE PROTEIN | | Descriptor: | BENZAMIDINE, MERCURY (II) ION, PROTEIN (METALLOCHAPERONE ATX1) | | Authors: | Rosenzweig, A.C, Huffman, D.L, Pufahl, M.Y.R.A, Hou, T.V.O, Wernimont, A.K. | | Deposit date: | 1999-03-04 | | Release date: | 1999-12-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Crystal structure of the Atx1 metallochaperone protein at 1.02 A resolution.
Structure Fold.Des., 7, 1999
|
|
1CC7
 
 | | CRYSTAL STRUCTURE OF THE ATX1 METALLOCHAPERONE PROTEIN | | Descriptor: | BENZAMIDINE, PROTEIN (METALLOCHAPERONE ATX1) | | Authors: | Rosenzweig, A.C, Huffman, D.L, Pufahl, M.Y.R.A, Hou, T.V.O, Wernimont, A.K. | | Deposit date: | 1999-03-04 | | Release date: | 1999-12-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of the Atx1 metallochaperone protein at 1.02 A resolution.
Structure Fold.Des., 7, 1999
|
|
1QE5
 
 | | PURINE NUCLEOSIDE PHOSPHORYLASE FROM CELLULOMONAS SP. IN COMPLEX WITH PHOSPHATE | | Descriptor: | CALCIUM ION, PENTOSYLTRANSFERASE, PHOSPHATE ION | | Authors: | Tebbe, J, Bzowska, A, Wielgus-Kutrowska, B, Schroeder, W, Kazimierczuk, Z, Shugar, D, Saenger, W, Koellner, G. | | Deposit date: | 1999-07-13 | | Release date: | 1999-12-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the purine nucleoside phosphorylase (PNP) from Cellulomonas sp. and its implication for the mechanism of trimeric PNPs.
J.Mol.Biol., 294, 1999
|
|
1QGV
 
 | | HUMAN SPLICEOSOMAL PROTEIN U5-15KD | | Descriptor: | SPLICEOSOMAL PROTEIN U5-15KD | | Authors: | Reuter, K, Nottrott, S, Fabrizio, P, Luehrmann, R, Ficner, R. | | Deposit date: | 1999-05-06 | | Release date: | 1999-12-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Identification, characterization and crystal structure analysis of the human spliceosomal U5 snRNP-specific 15 kD protein.
J.Mol.Biol., 294, 1999
|
|
1QOV
 
 | | PHOTOSYNTHETIC REACTION CENTER MUTANT WITH ALA M260 REPLACED WITH TRP (CHAIN M, A260W) | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CARDIOLIPIN, ... | | Authors: | McAuley, K.E, Fyfe, P.K, Ridge, J.P, Isaacs, N.W, Cogdell, R.J, Jones, M.R. | | Deposit date: | 1999-11-17 | | Release date: | 1999-12-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Details of an Interaction between Cardiolipin and an Integral Membrane Protein
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
2CRX
 
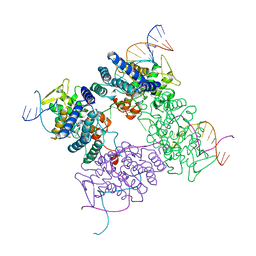 | |
1QMC
 
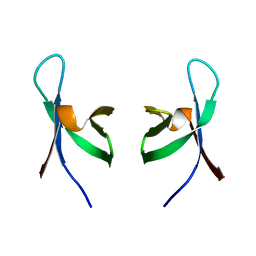 | | C-terminal DNA-binding domain of HIV-1 integrase, NMR, 42 structures | | Descriptor: | HIV-1 INTEGRASE | | Authors: | Eijkelenboom, A.P.A.M, Sprangers, R, Hard, K, Puras Lutzke, R.A, Plasterk, R.H.A, Boelens, R, Kaptein, R. | | Deposit date: | 1999-09-27 | | Release date: | 1999-12-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Refined Solution Structure of the C-Terminal DNA-Binding Domain of Human Immunovirus-1 Integrase.
Proteins: Struct.,Funct., Genet., 36, 1999
|
|
3CRX
 
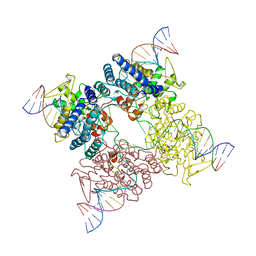 | |
1QMZ
 
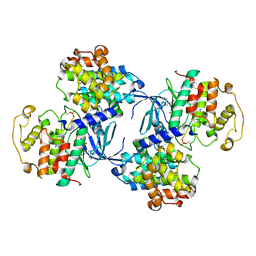 | | PHOSPHORYLATED CDK2-CYCLYIN A-SUBSTRATE PEPTIDE COMPLEX | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CELL DIVISION PROTEIN KINASE 2, G2/MITOTIC-SPECIFIC CYCLIN A, ... | | Authors: | Brown, N.R, Noble, M.E.M, Endicott, J.A, Johnson, L.N. | | Deposit date: | 1999-10-11 | | Release date: | 1999-12-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structural Basis for Specificity of Substrate and Recruitment Peptides for Cyclin-Dependent Kinases
Nat.Cell Biol., 1, 1999
|
|
1QL6
 
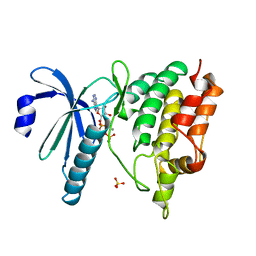 | | THE CATALYTIC MECHANISM OF PHOSPHORYLASE KINASE PROBED BY MUTATIONAL STUDIES | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, PHOSPHORYLASE KINASE, ... | | Authors: | Skamnaki, V.T, Owen, D.J, Noble, M.E.M, Lowe, E.D, Oikonomakos, N.G, Johnson, L.N. | | Deposit date: | 1999-08-24 | | Release date: | 1999-12-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Catalytic Mechanism of Phosphorylase Kinase Probed by Mutational Studies.
Biochemistry, 38, 1999
|
|
2KZZ
 
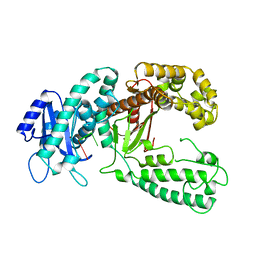 | | KLENOW FRAGMENT WITH NORMAL SUBSTRATE AND ZINC ONLY | | Descriptor: | DNA (5'-D(*GP*CP*TP*T*AP*CP*G)-3'), PROTEIN (DNA POLYMERASE I), ZINC ION | | Authors: | Brautigam, C.A, Sun, S, Piccirilli, J.A, Steitz, T.A. | | Deposit date: | 1998-07-07 | | Release date: | 1999-12-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structures of normal single-stranded DNA and deoxyribo-3'-S-phosphorothiolates bound to the 3'-5' exonucleolytic active site of DNA polymerase I from Escherichia coli.
Biochemistry, 38, 1999
|
|
1R1B
 
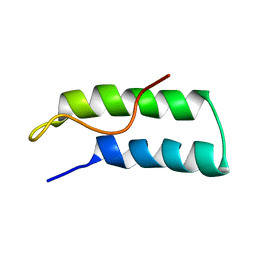 | | EPRS SECOND REPEATED ELEMENT, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | TRNA SYNTHETASE | | Authors: | Cahuzac, B, Berthonneau, E, Birlirakis, N, Mirande, M, Guittet, E. | | Deposit date: | 1998-12-15 | | Release date: | 1999-12-15 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | A recurrent RNA-binding domain is appended to eukaryotic aminoacyl-tRNA synthetases.
EMBO J., 19, 2000
|
|
2PFL
 
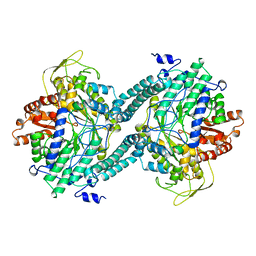 | | CRYSTAL STRUCTURE OF PFL FROM E.COLI | | Descriptor: | CHLORIDE ION, PROTEIN (PYRUVATE FORMATE-LYASE), SODIUM ION | | Authors: | Becker, A, Fritz-Wolf, K, Kabsch, W, Knappe, J, Schultz, S, Wagner, A.F.V. | | Deposit date: | 1999-05-26 | | Release date: | 1999-12-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure and mechanism of the glycyl radical enzyme pyruvate formate-lyase.
Nat.Struct.Biol., 6, 1999
|
|
1CX6
 
 | |
3EZB
 
 | | COMPLEX OF THE AMINO TERMINAL DOMAIN OF ENZYME I AND THE HISTIDINE-CONTAINING PHOSPHOCARRIER PROTEIN HPR FROM ESCHERICHIA COLI | | Descriptor: | PROTEIN (PHOSPHOCARRIER PROTEIN HPR), PROTEIN (PHOSPHOTRANSFER SYSTEM, ENZYME I) | | Authors: | Clore, G.M, Garrett, D.S, Gronenborn, A.M. | | Deposit date: | 1998-11-03 | | Release date: | 1999-12-16 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the 40,000 Mr phosphoryl transfer complex between the N-terminal domain of enzyme I and HPr.
Nat.Struct.Biol., 6, 1999
|
|
1QM1
 
 | |
1QM2
 
 | |
1QM3
 
 | |
1QLZ
 
 | | Human prion protein | | Descriptor: | PRION PROTEIN | | Authors: | Zahn, R, Liu, A, Luhrs, T, Wuthrich, K. | | Deposit date: | 1999-09-20 | | Release date: | 1999-12-16 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of the Human Prion Protein
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
