4M99
 
 | |
3QDH
 
 | |
5OWB
 
 | |
5OWE
 
 | |
5OWK
 
 | |
5E73
 
 | | Crystal Structure of BAZ2B bromodomain in complex with acetylindole compound UZH47 | | Descriptor: | Bromodomain adjacent to zinc finger domain protein 2B, N-(1-acetyl-1H-indol-3-yl)-N-(5-hydroxy-2-methylphenyl)acetamide | | Authors: | Lolli, G, Spiliotopoulos, D, Unzue, A, Nevado, C, Caflisch, A. | | Deposit date: | 2015-10-11 | | Release date: | 2015-10-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | The "Gatekeeper" Residue Influences the Mode of Binding of Acetyl Indoles to Bromodomains.
J. Med. Chem., 59, 2016
|
|
5E74
 
 | | Crystal Structure of BAZ2B bromodomain in complex with acetylindole compound UZH50 | | Descriptor: | Bromodomain adjacent to zinc finger domain protein 2B, N-(1-acetyl-1H-indol-3-yl)-N-(5-hydroxy-2-methylphenyl)-3-(trifluoromethyl)benzamide | | Authors: | Lolli, G, Spiliotopoulos, D, Dolbois, A, Nevado, C, Caflisch, A. | | Deposit date: | 2015-10-11 | | Release date: | 2015-10-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.783 Å) | | Cite: | The "Gatekeeper" Residue Influences the Mode of Binding of Acetyl Indoles to Bromodomains.
J. Med. Chem., 59, 2016
|
|
1QHD
 
 | | CRYSTAL STRUCTURE OF VP6, THE MAJOR CAPSID PROTEIN OF GROUP A ROTAVIRUS | | Descriptor: | CALCIUM ION, CHLORIDE ION, VIRAL CAPSID VP6, ... | | Authors: | Mathieu, M, Petitpas, I, Rey, F.A. | | Deposit date: | 1999-04-29 | | Release date: | 2001-04-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Atomic structure of the major capsid protein of rotavirus: implications for the architecture of the virion.
EMBO J., 20, 2001
|
|
2E6K
 
 | | X-ray structure of Thermus thermopilus HB8 TT0505 | | Descriptor: | Transketolase | | Authors: | Yoshida, H, Kamitori, S, Agari, Y, Iino, H, Kanagawa, M, Nakagawa, N, Ebihara, A, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-12-27 | | Release date: | 2007-11-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | X-ray structure of Thermus thermophilus HB8 TT0505
To be Published
|
|
6LCJ
 
 | |
1PAK
 
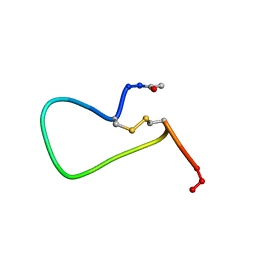 | | NMR SOLUTION STRUCTURE AND FLEXIBILITY OF A PEPTIDE ANTIGEN REPRESENTING THE RECEPTOR BINDING DOMAIN OF PSEUDOMONAS AERUGINOSA | | Descriptor: | FIMBRIAL PROTEIN PRECURSOR, HYDROXIDE ION | | Authors: | Mcinnes, C, Sonnichsen, F.D, Kay, C.M, Hodges, R.S, Sykes, B.D. | | Deposit date: | 1993-08-25 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure and flexibility of a peptide antigen representing the receptor binding domain of Pseudomonas aeruginosa.
Biochemistry, 32, 1993
|
|
1PAJ
 
 | | NMR SOLUTION STRUCTURE AND FLEXIBILITY OF A PEPTIDE ANTIGEN REPRESENTING THE RECEPTOR BINDING DOMAIN OF PSEUDOMONAS AERUGINOSA | | Descriptor: | FIMBRIAL PROTEIN PRECURSOR, HYDROXIDE ION | | Authors: | Mcinnes, C, Sonnichsen, F.D, Kay, C.M, Hodges, R.S, Sykes, B.D. | | Deposit date: | 1993-08-25 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure and flexibility of a peptide antigen representing the receptor binding domain of Pseudomonas aeruginosa.
Biochemistry, 32, 1993
|
|
8CO1
 
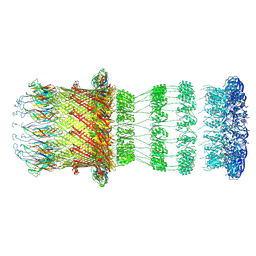 | | Type II Secretion System | | Descriptor: | IPT/TIG domain-containing protein, Lipoprotein, Probable type IV piliation system protein DR_0774 | | Authors: | Farci, D, Piano, D. | | Deposit date: | 2023-02-26 | | Release date: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Structural characterization and functional insights into the type II secretion system of the poly-extremophile Deinococcus radiodurans.
J.Biol.Chem., 300, 2024
|
|
7PNB
 
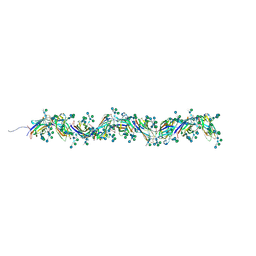 | | Sulfolobus acidocaldarius 0406 filament. | | Descriptor: | 6-deoxy-6-sulfo-beta-D-glucopyranose-(1-3)-[alpha-D-mannopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Sulfolobus acidocaldarius 0406 filament., beta-D-glucopyranose-(1-4)-6-deoxy-6-sulfo-beta-D-glucopyranose-(1-3)-[alpha-D-mannopyranose-(1-4)][alpha-D-mannopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Isupov, M.N, Gaines, M, Daum, B. | | Deposit date: | 2021-09-06 | | Release date: | 2022-09-14 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Electron cryo-microscopy reveals the structure of the archaeal thread filament.
Nat Commun, 13, 2022
|
|
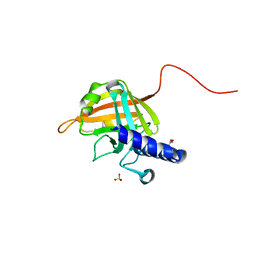
7D6P
 
 | |
7D6T
 
 | |
5NWP
 
 | |
5VQ5
 
 | | Crystal Structure of the Lectin Domain From the F17-like Adhesin, UclD | | Descriptor: | Adhesin, IODIDE ION | | Authors: | Klein, R.D, Spaulding, C.N, Dodson, K.W, Pinkner, J.S, Hultgren, S.J, Fremont, D. | | Deposit date: | 2017-05-08 | | Release date: | 2017-05-24 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Selective depletion of uropathogenic E. coli from the gut by a FimH antagonist.
Nature, 546, 2017
|
|
4B0M
 
 | | Complex of the Caf1AN usher domain, Caf1M chaperone and Caf1 subunit from Yersinia pestis | | Descriptor: | CHAPERONE PROTEIN CAF1M, F1 CAPSULE ANTIGEN, F1 CAPSULE-ANCHORING PROTEIN | | Authors: | Dubnovitsky, A, Yu, X.D, Pudney, A.F, MacIntyre, S, Knight, S.D, Zavialov, A.V. | | Deposit date: | 2012-07-03 | | Release date: | 2012-09-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Allosteric Mechanism Controls Traffic in the Chaperone/Usher Pathway.
Structure, 20, 2012
|
|
7O3V
 
 | | Stalk complex structure (TrwJ/VirB5-TrwI/VirB6) from the fully-assembled R388 type IV secretion system determined by cryo-EM. | | Descriptor: | TrwI protein, TrwJ protein | | Authors: | Mace, K, Vadakkepat, A.K, Lukoyanova, N, Waksman, G. | | Deposit date: | 2021-04-03 | | Release date: | 2022-06-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of a type IV secretion system.
Nature, 607, 2022
|
|
7O43
 
 | |
7O3J
 
 | | O-layer structure (TrwH/VirB7, TrwF/VirB9CTD, TrwE/VirB10CTD) of the outer membrane core complex from the fully-assembled R388 type IV secretion system determined by cryo-EM. | | Descriptor: | TrwE protein, TrwF protein, TrwH protein | | Authors: | Mace, K, Vadakkepat, A.K, Lukoyanova, N, Waksman, G. | | Deposit date: | 2021-04-01 | | Release date: | 2022-06-22 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Cryo-EM structure of a type IV secretion system.
Nature, 607, 2022
|
|
7O3T
 
 | | I-layer structure (TrwF/VirB9NTD, TrwE/VirB10NTD) of the outer membrane core complex from the fully-assembled R388 type IV secretion system determined by cryo-EM. | | Descriptor: | TrwE protein, TrwF protein | | Authors: | Mace, K, Vadakkepat, A.K, Lukoyanova, N, Waksman, G. | | Deposit date: | 2021-04-03 | | Release date: | 2022-06-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure of a type IV secretion system.
Nature, 607, 2022
|
|
7O42
 
 | | TrwK/VirB4unbound trimer of dimers complex (with Hcp1) from the R388 type IV secretion system determined by cryo-EM. | | Descriptor: | TrwK protein,Protein hcp1 | | Authors: | Vadakkepat, A.K, Mace, K, Lukoyanova, N, Waksman, G. | | Deposit date: | 2021-04-04 | | Release date: | 2022-06-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structure of a type IV secretion system.
Nature, 607, 2022
|
|
7O41
 
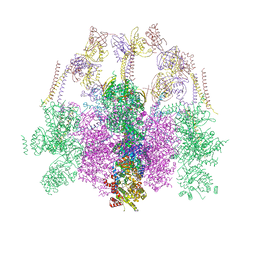 | | Hexameric composite model of the Inner Membrane Complex (IMC) with the Arches from the fully-assembled R388 type IV secretion system determined by cryo-EM. | | Descriptor: | TrwG protein, TrwK protein, TrwM protein | | Authors: | Mace, K, Vadakkepat, A.K, Lukoyanova, N, Waksman, G. | | Deposit date: | 2021-04-04 | | Release date: | 2022-06-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Cryo-EM structure of a type IV secretion system.
Nature, 607, 2022
|
|
