8CWA
 
 | |
4WC8
 
 | |
7UBF
 
 | |
7UBI
 
 | |
7UBE
 
 | |
8SZR
 
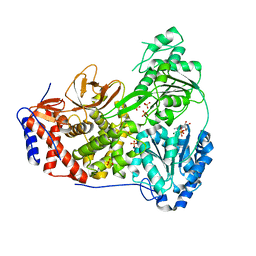 | | Dog DHX9 bound to ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, RNA helicase, ... | | Authors: | Lee, Y.-T, Sickmier, E.A, Grigoriu, S, Boriack-Sjodin, P.A. | | Deposit date: | 2023-05-30 | | Release date: | 2023-08-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Crystal structures of the DExH-box RNA helicase DHX9.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8SZQ
 
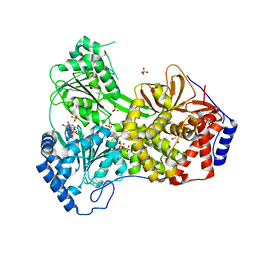 | | Cat DHX9 bound to ADP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Lee, Y.-T, Sickmier, E.A, Grigoriu, S, Boriack-Sjodin, P.A. | | Deposit date: | 2023-05-30 | | Release date: | 2023-08-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.711 Å) | | Cite: | Crystal structures of the DExH-box RNA helicase DHX9.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8SZP
 
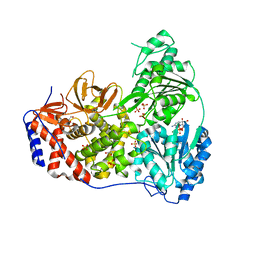 | | Human DHX9 bound to ADP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase A, ... | | Authors: | Lee, Y.-T, Sickmier, E.A, Grigoriu, S, Boriack-Sjodin, P.A. | | Deposit date: | 2023-05-30 | | Release date: | 2023-08-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Crystal structures of the DExH-box RNA helicase DHX9.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7JRH
 
 | |
5W5U
 
 | |
5W6T
 
 | |
7UZL
 
 | |
7UBC
 
 | |
7UBG
 
 | |
8UN5
 
 | | KRAS-G13D-GDP in complex with Cpd38 ((E)-1-((3S)-4-(7-(6-amino-4-methyl-3-(trifluoromethyl)pyridin-2-yl)-6-chloro-8-fluoro-2-(((S)-2-methylenetetrahydro-1H-pyrrolizin-7a(5H)-yl)methoxy)quinazolin-4-yl)-3-methylpiperazin-1-yl)-3-(1,2,3,4-tetrahydroisoquinolin-8-yl)prop-2-en-1-one) | | Descriptor: | (2E)-1-{(3S)-4-[(7M)-7-[6-amino-4-methyl-3-(trifluoromethyl)pyridin-2-yl]-6-chloro-8-fluoro-2-{[(4R,7aS)-2-methylidenetetrahydro-1H-pyrrolizin-7a(5H)-yl]methoxy}quinazolin-4-yl]-3-methylpiperazin-1-yl}-3-(1,2,3,4-tetrahydroisoquinolin-8-yl)prop-2-en-1-one, GLYCEROL, GTPase KRas, ... | | Authors: | Ultsch, M.H. | | Deposit date: | 2023-10-18 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Structure-Based Design and Evaluation of Reversible KRAS G13D Inhibitors.
Acs Med.Chem.Lett., 15, 2024
|
|
8AV2
 
 | | Crystal structure for the FnIII module of mouse LEP-R in complex with the anti-LEP-R nanobody VHH-4.80 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Leptin receptor, SULFATE ION, ... | | Authors: | Verstraete, K, Savvides, S.N, Verschueren, K.G, Tsirigotaki, A. | | Deposit date: | 2022-08-26 | | Release date: | 2023-04-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Mechanism of receptor assembly via the pleiotropic adipokine Leptin.
Nat.Struct.Mol.Biol., 30, 2023
|
|
3QXC
 
 | | Crystal structure of dethiobiotin synthetase (BioD) from Helicobacter pylori complexed with ATP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Porebski, P.J, Klimecka, M.M, Chruszcz, M, Murzyn, K, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-03-01 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Structural characterization of Helicobacter pylori dethiobiotin synthetase reveals differences between family members.
Febs J., 279, 2012
|
|
3QXJ
 
 | | Crystal structure of dethiobiotin synthetase (BioD) from Helicobacter pylori complexed with GTP | | Descriptor: | 1,2-ETHANEDIOL, Dethiobiotin synthetase, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Klimecka, M.M, Porebski, P.J, Chruszcz, M, Murzyn, K, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-03-01 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Structural characterization of Helicobacter pylori dethiobiotin synthetase reveals differences between family members.
Febs J., 279, 2012
|
|
3QY0
 
 | | Crystal structure of dethiobiotin synthetase (BioD) from Helicobacter pylori complexed with GDP | | Descriptor: | 1,2-ETHANEDIOL, Dethiobiotin synthetase, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Porebski, P.J, Klimecka, M.M, Chruszcz, M, Murzyn, K, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-03-02 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural characterization of Helicobacter pylori dethiobiotin synthetase reveals differences between family members.
Febs J., 279, 2012
|
|
3QXH
 
 | | Crystal structure of dethiobiotin synthetase (BioD) from Helicobacter pylori complexed with ADP and 8-aminocaprylic acid | | Descriptor: | 1,2-ETHANEDIOL, 8-aminooctanoic acid, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Porebski, P.J, Klimecka, M.M, Chruszcz, M, Murzyn, K, Minor, C, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-03-01 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structural characterization of Helicobacter pylori dethiobiotin synthetase reveals differences between family members.
Febs J., 279, 2012
|
|
3QXS
 
 | | Crystal structure of dethiobiotin synthetase (BioD) from Helicobacter pylori complexed with ANP | | Descriptor: | 1,2-ETHANEDIOL, Dethiobiotin synthetase, MAGNESIUM ION, ... | | Authors: | Klimecka, M.M, Porebski, P.J, Chruszcz, M, Jablonska, K, Murzyn, K, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-03-02 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural characterization of Helicobacter pylori dethiobiotin synthetase reveals differences between family members.
Febs J., 279, 2012
|
|
3QXX
 
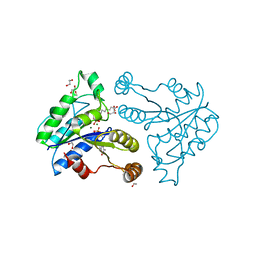 | | Crystal structure of dethiobiotin synthetase (BioD) from Helicobacter pylori complexed with GDP and 8-aminocaprylic acid | | Descriptor: | 1,2-ETHANEDIOL, 8-aminooctanoic acid, Dethiobiotin synthetase, ... | | Authors: | Porebski, P.J, Klimecka, M.M, Chruszcz, M, Murzyn, K, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-03-02 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structural characterization of Helicobacter pylori dethiobiotin synthetase reveals differences between family members.
Febs J., 279, 2012
|
|
1M8Q
 
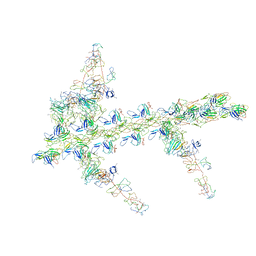 | | Molecular Models of Averaged Rigor Crossbridges from Tomograms of Insect Flight Muscle | | Descriptor: | Skeletal muscle Actin, Skeletal muscle Myosin II, Skeletal muscle Myosin II Essential Light Chain, ... | | Authors: | Chen, L.F, Winkler, H, Reedy, M.K, Reedy, M.C, Taylor, K.A. | | Deposit date: | 2002-07-25 | | Release date: | 2002-09-10 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (70 Å) | | Cite: | Molecular modeling of averaged rigor crossbridges from tomograms of insect flight muscle.
J.Struct.Biol., 138, 2002
|
|
2IMQ
 
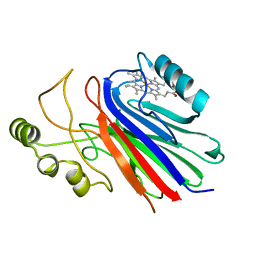 | | Crystal structure of ferrous cimex nitrophorin | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Salivary nitrophorin | | Authors: | Weichsel, A, Montfort, W.R. | | Deposit date: | 2006-10-04 | | Release date: | 2007-08-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystallographic and spectroscopic characterization of ferrous complexes of cimex nitrophorin, a heme-thiolate protein from the bedbug
To be Published
|
|
5Z1R
 
 | | Crystal Structure Analysis of the BRD4 | | Descriptor: | 1,2-ETHANEDIOL, 5-bromo-N-(2,2-dimethyl-3-oxo-3,4-dihydro-2H-1,4-benzoxazin-7-yl)-2-methoxybenzene-1-sulfonamide, Bromodomain-containing protein 4, ... | | Authors: | Xu, Y, Zhang, Y, Xiang, Q, Song, M, Wang, C. | | Deposit date: | 2017-12-28 | | Release date: | 2019-01-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Y08060: A Selective BET Inhibitor for Treatment of Prostate Cancer.
Acs Med.Chem.Lett., 9, 2018
|
|
