6QUG
 
 | | GHK tagged MBP-Nup98(1-29) | | Descriptor: | COPPER (II) ION, Maltodextrin-binding protein,Nucleoporin, putative, ... | | Authors: | Huyton, T, Gorlich, D. | | Deposit date: | 2019-02-27 | | Release date: | 2020-05-27 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The copper(II)-binding tripeptide GHK, a valuable crystallization and phasing tag for macromolecular crystallography.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6U1D
 
 | | Thermus thermophilus D-alanine-D-alanine ligase in complex with ATP, D-alanine-D-alanine, Mg2+ and Rb+ | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, D-ALANINE, D-alanine--D-alanine ligase, ... | | Authors: | Pederick, J.L, Bruning, J.B. | | Deposit date: | 2019-08-15 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | d-Alanine-d-alanine ligase as a model for the activation of ATP-grasp enzymes by monovalent cations.
J.Biol.Chem., 295, 2020
|
|
6U1J
 
 | | Thermus thermophilus D-alanine-D-alanine ligase in complex with ADP, phosphate, D-ala-D-ala, Mg2+ and K+ | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, D-ALANINE, D-alanine--D-alanine ligase, ... | | Authors: | Pederick, J.L, Bruning, J.B, Thompson, A.P. | | Deposit date: | 2019-08-15 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | d-Alanine-d-alanine ligase as a model for the activation of ATP-grasp enzymes by monovalent cations.
J.Biol.Chem., 295, 2020
|
|
5UU8
 
 | |
6U1E
 
 | | Thermus thermophilus D-alanine-D-alanine ligase in complex with ATP, D-alanine-D-alanine, Mg2+ and Rb+ | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, D-ALANINE, D-alanine--D-alanine ligase, ... | | Authors: | Pederick, J.L, Bruning, J.B. | | Deposit date: | 2019-08-15 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | d-Alanine-d-alanine ligase as a model for the activation of ATP-grasp enzymes by monovalent cations.
J.Biol.Chem., 295, 2020
|
|
1M77
 
 | | Near Atomic Resolution Crystal Structure of an A-DNA Decamer d(CCCGATCGGG): Cobalt Hexammine Interactions with A-DNA | | Descriptor: | 5'-D(*CP*CP*CP*GP*AP*TP*CP*GP*GP*G)-3', COBALT HEXAMMINE(III) | | Authors: | Ramakrishnan, B, Sekharudu, C, Pan, B.C, Sundaralingam, M. | | Deposit date: | 2002-07-18 | | Release date: | 2003-01-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Near-atomic resolution crystal structure of an A-DNA decamer d(CCCGATCGGG): cobalt hexammine interaction with A-DNA.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
6U1F
 
 | | Thermus thermophilus D-alanine-D-alanine ligase in complex with ATP, D-alanine-D-alanine, Mg2+ and Cs+ | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CESIUM ION, D-ALANINE, ... | | Authors: | Pederick, J.L, Bruning, J.B. | | Deposit date: | 2019-08-15 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | d-Alanine-d-alanine ligase as a model for the activation of ATP-grasp enzymes by monovalent cations.
J.Biol.Chem., 295, 2020
|
|
4XIS
 
 | |
4ZKK
 
 | | The novel double-fold structure of d(GCATGCATGC) | | Descriptor: | COBALT (II) ION, DNA (5'-D(*GP*CP*AP*TP*GP*CP*AP*TP*GP*C)-3') | | Authors: | Thirugnanasambandam, A, Karthik, S, Mandal, P.K, Gautham, N. | | Deposit date: | 2015-04-30 | | Release date: | 2015-10-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | The novel double-folded structure of d(GCATGCATGC): a possible model for triplet-repeat sequences
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7CHT
 
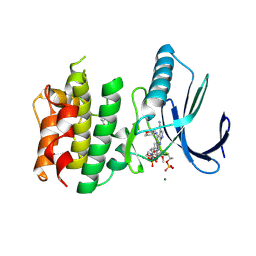 | | Crystal structure of TTK kinase domain in complex with compound 30 | | Descriptor: | 2-[[2-methoxy-4-(2-oxidanylidenepyrrolidin-1-yl)phenyl]amino]-4-(oxan-4-ylamino)-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, Dual specificity protein kinase TTK, MAGNESIUM ION | | Authors: | Kim, H.L, Cho, H.Y, Park, Y.W, Lee, Y.H, Ko, E.H, Choi, H.G, Son, J.B, Kim, N.D. | | Deposit date: | 2020-07-06 | | Release date: | 2021-05-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray Crystal Structure-Guided Design and Optimization of 7 H -Pyrrolo[2,3- d ]pyrimidine-5-carbonitrile Scaffold as a Potent and Orally Active Monopolar Spindle 1 Inhibitor.
J.Med.Chem., 64, 2021
|
|
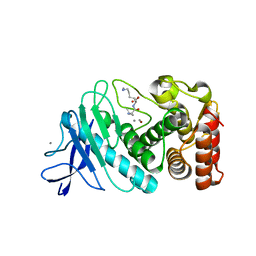
6D5R
 
 | |
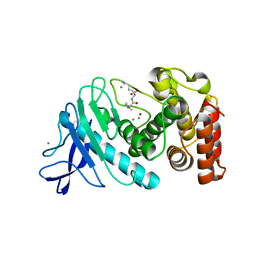
6D5Q
 
 | |
6D5N
 
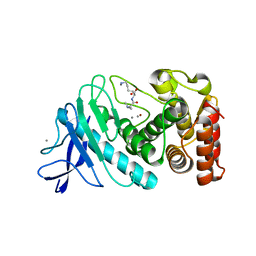 | | Hexagonal thermolysin (295) in the presence of 50% xylose | | Descriptor: | CALCIUM ION, LYSINE, Thermolysin, ... | | Authors: | Juers, D.H. | | Deposit date: | 2018-04-19 | | Release date: | 2018-10-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.00003672 Å) | | Cite: | The impact of cryosolution thermal contraction on proteins and protein crystals: volumes, conformation and order.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5UUB
 
 | |
6D6F
 
 | |
6D5P
 
 | |
1O0D
 
 | | Human Thrombin complexed with a d-Phe-Pro-Arg-type Inhibitor and a C-terminal Hirudin derived exo-site inhibitor | | Descriptor: | (2-{2-[(5-CARBAMIMIDOYL-1-METHYL-1H-PYRROL-2-YLMETHYL)-CARBAMOYL]-PYRROL-1-YL}- 1-CYCLOHEXYLMETHYL-2-OXO-ETHYLAMINO)-ACETIC ACID, Decapeptide Hirudin Analogue, Thrombin heavy chain, ... | | Authors: | Lange, U.E, Bauke, D, Hornberger, W, Mack, H, Seitz, W, Hoeffken, H.W. | | Deposit date: | 2003-02-21 | | Release date: | 2003-10-14 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | D-Phe-Pro-Arg type thrombin inhibitors: unexpected selectivity by modification of the P1 moiety
Bioorg.Med.Chem.Lett., 13, 2003
|
|
6JMX
 
 | | Structure of open form of peptidoglycan peptidase | | Descriptor: | D(-)-TARTARIC ACID, GLYCEROL, Peptidase M23, ... | | Authors: | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | Deposit date: | 2019-03-13 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.859 Å) | | Cite: | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
5EUV
 
 | | Crystal structure of a cold-adapted dimeric beta-D-galactosidase from Paracoccus sp. 32d strain | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, Beta-D-galactosidase, ... | | Authors: | Rutkiewicz-Krotewicz, M, Bujacz, A, Pietrzyk, A.J, Sekula, B, Bujacz, G. | | Deposit date: | 2015-11-19 | | Release date: | 2016-09-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural studies of a cold-adapted dimeric beta-D-galactosidase from Paracoccus sp. 32d.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
6JN7
 
 | | Structure of H216A mutant closed form peptidoglycan peptidase | | Descriptor: | D(-)-TARTARIC ACID, Peptidase M23, ZINC ION | | Authors: | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | Deposit date: | 2019-03-13 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
6AGL
 
 | |
3E8M
 
 | | Structure-function Analysis of 2-Keto-3-deoxy-D-glycero-D-galacto-nononate-9-phosphate (KDN) Phosphatase Defines a New Clad Within the Type C0 HAD Subfamily | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, Acylneuraminate cytidylyltransferase, ... | | Authors: | Lu, Z, Wang, L, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2008-08-20 | | Release date: | 2008-11-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structure-Function Analysis of 2-Keto-3-deoxy-D-glycero-D-galactonononate-9-phosphate Phosphatase Defines Specificity Elements in Type C0 Haloalkanoate Dehalogenase Family Members.
J.Biol.Chem., 284, 2009
|
|
6U1G
 
 | | Thermus thermophilus D-alanine-D-alanine ligase in complex with ATP, D-alanine-D-alanine, Mg2+ and Cs+ | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CESIUM ION, D-ALANINE, ... | | Authors: | Pederick, J.L, Bruning, J.B. | | Deposit date: | 2019-08-15 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | d-Alanine-d-alanine ligase as a model for the activation of ATP-grasp enzymes by monovalent cations.
J.Biol.Chem., 295, 2020
|
|
6DZF
 
 | |
3IRX
 
 | | Crystal Structure of HIV-1 reverse transcriptase (RT) in complex with the Non-nucleoside RT Inhibitor (E)-S-Methyl 5-(1-(3,7-Dimethyl-2-oxo-2,3-dihydrobenzo[d]oxazol-5-yl)-5-(5-methyl-1,3,4-oxadiazol-2-yl)pent-1-enyl)-2-methoxy-3-methylbenzothioate. | | Descriptor: | (E)-S-Methyl 5-(1-(3,7-Dimethyl-2-oxo-2,3-dihydrobenzo[d]oxazol-5-yl)-5-(5-methyl-1,3,4-oxadiazol-2-yl)pent-1-enyl)-2-methoxy-3-methy lbenzothioate, Reverse transcriptase, Reverse transcriptase/ribonuclease H | | Authors: | Ho, W.C, Arnold, E. | | Deposit date: | 2009-08-24 | | Release date: | 2010-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of HIV-1 reverse transcriptase (RT) in complex with the
alkenyldiarylmethane (ADAM) Non-nucleoside RT Inhibitor (E)-S-Methyl
5-(1-(3,7-Dimethyl-2-oxo-2,3-dihydrobenzo[d]oxazol-5-yl)-5-(5-methyl-1,3,4-oxadiazol-2-yl)pent-1-enyl)-2-methoxy-3-methylbenzothioate.
J.Med.Chem., 52, 2009
|
|
