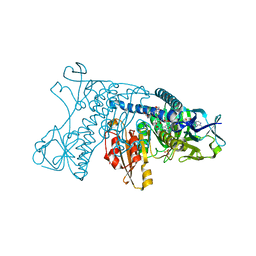
1GRE
 
 | |
1GRF
 
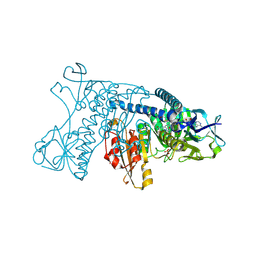 | |
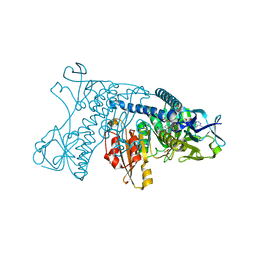
1GRG
 
 | |
1GRH
 
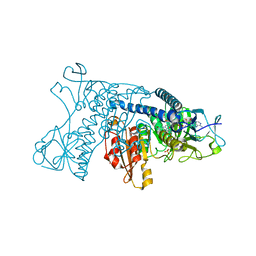 | | INHIBITION OF HUMAN GLUTATHIONE REDUCTASE BY THE NITROSOUREA DRUGS 1,3-BIS(2-CHLOROETHYL)-1-NITROSOUREA AND 1-(2-CHLOROETHYL)-3-(2-HYDROXYETHYL)-1-NITROSOUREA | | Descriptor: | ETHANOL, FLAVIN-ADENINE DINUCLEOTIDE, GLUTATHIONE REDUCTASE, ... | | Authors: | Karplus, P.A, Schulz, G.E. | | Deposit date: | 1992-12-15 | | Release date: | 1994-01-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Inhibition of human glutathione reductase by the nitrosourea drugs 1,3-bis(2-chloroethyl)-1-nitrosourea and 1-(2-chloroethyl)-3-(2-hydroxyethyl)-1-nitrosourea. A crystallographic analysis.
Eur.J.Biochem., 171, 1988
|
|
1GRI
 
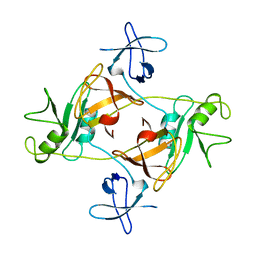 | | GRB2 | | Descriptor: | GROWTH FACTOR BOUND PROTEIN 2 | | Authors: | Maignan, S, Arnoux, B, Ducruix, A. | | Deposit date: | 1995-02-28 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of the mammalian Grb2 adaptor.
Science, 268, 1995
|
|
1GRJ
 
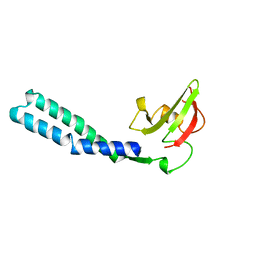 | |
1GRL
 
 | | THE CRYSTAL STRUCTURE OF THE BACTERIAL CHAPERONIN GROEL AT 2.8 ANGSTROMS | | Descriptor: | GROEL (HSP60 CLASS) | | Authors: | Braig, K, Otwinowski, Z, Hegde, R, Boisvert, D.C, Joachimiak, A, Horwich, A.L, Sigler, P.B. | | Deposit date: | 1995-03-07 | | Release date: | 1995-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of the bacterial chaperonin GroEL at 2.8 A.
Nature, 371, 1994
|
|
1GRM
 
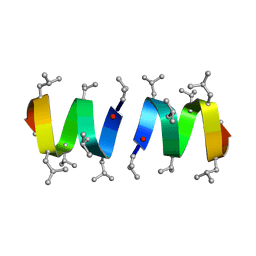 | | REFINEMENT OF THE SPATIAL STRUCTURE OF THE GRAMICIDIN A TRANSMEMBRANE ION-CHANNEL (RUSSIAN) | | Descriptor: | GRAMICIDIN A | | Authors: | Arseniev, A.S, Barsukov, I.L, Lomize, A.L, Orekhov, V.Y, Bystrov, V.F. | | Deposit date: | 1993-10-18 | | Release date: | 1994-01-31 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Refinement of the Spatial Structure of the Gramicidin a Ion Channel
Biol.Membr.(Ussr), 18, 1992
|
|
1GRN
 
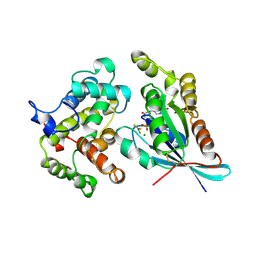 | | CRYSTAL STRUCTURE OF THE CDC42/CDC42GAP/ALF3 COMPLEX. | | Descriptor: | ALUMINUM FLUORIDE, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Nassar, N, Hoffman, G.R, Clardy, J.C, Cerione, R.A. | | Deposit date: | 1998-07-30 | | Release date: | 1999-12-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of Cdc42 bound to the active and catalytically compromised forms of Cdc42GAP.
Nat.Struct.Biol., 5, 1998
|
|
1GRO
 
 | |
1GRP
 
 | |
1GRQ
 
 | |
1GRR
 
 | |
1GRT
 
 | | HUMAN GLUTATHIONE REDUCTASE A34E/R37W MUTANT | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLUTATHIONE REDUCTASE | | Authors: | Stoll, V.S, Simpson, S.J, Krauth-Siegel, R.L, Walsh, C.T, Pai, E.F. | | Deposit date: | 1996-12-17 | | Release date: | 1997-06-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Glutathione reductase turned into trypanothione reductase: structural analysis of an engineered change in substrate specificity.
Biochemistry, 36, 1997
|
|
1GRU
 
 | | SOLUTION STRUCTURE OF GROES-ADP7-GROEL-ATP7 COMPLEX BY CRYO-EM | | Descriptor: | GROEL, GROES | | Authors: | Ranson, N.A, Farr, G.W, Roseman, A.M, Gowen, B, Fenton, W.A, Horwich, A.L, Saibil, H.R. | | Deposit date: | 2001-12-16 | | Release date: | 2002-01-28 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (12.5 Å) | | Cite: | ATP-Bound States of Groel Captured by Cryo-Electron Microscopy.
Cell(Cambridge,Mass.), 107, 2001
|
|
1GRV
 
 | | Hypoxanthine Phosphoribosyltransferase from E. coli | | Descriptor: | HYPOXANTHINE PHOSPHORIBOSYLTRANSFERASE, MAGNESIUM ION | | Authors: | Guddat, L.W, Vos, S, Martin, J.L, Keough, D.T, De Jersey, J. | | Deposit date: | 2001-12-17 | | Release date: | 2002-12-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structures of Free, Imp-, and Gmp- Bound Escherichia Coli Hypoxanthine Phosphoribosyltransferase
Protein Sci., 11, 2002
|
|
1GRW
 
 | | C. elegans major sperm protein | | Descriptor: | MAJOR SPERM PROTEIN 31/40/142 | | Authors: | Baker, A.M.E, Roberts, T.M, Stewart, M. | | Deposit date: | 2001-12-18 | | Release date: | 2002-06-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 2.6 A Resolution Crystal Structure of Helices of the Motile Major Sperm Protein (Msp) of Caenorhabditis Elegans
J.Mol.Biol., 319, 2002
|
|
1GRX
 
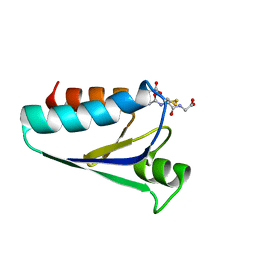 | | STRUCTURE OF E. COLI GLUTAREDOXIN | | Descriptor: | GLUTAREDOXIN, GLUTATHIONE | | Authors: | Bushweller, J.H, Billeter, M, Holmgren, L.A, Wuthrich, K. | | Deposit date: | 1993-10-01 | | Release date: | 1994-01-31 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | NMR structure of oxidized Escherichia coli glutaredoxin: comparison with reduced E. coli glutaredoxin and functionally related proteins.
Protein Sci., 1, 1992
|
|
1GRZ
 
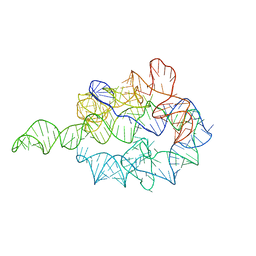 | |
1GS0
 
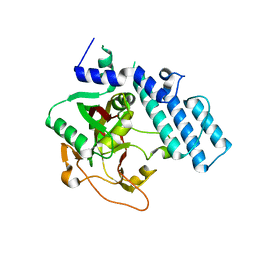 | |
1GS3
 
 | |
1GS4
 
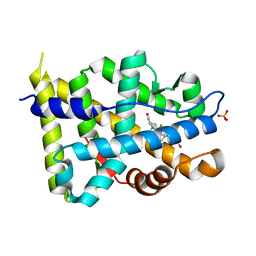 | | Structural basis for the glucocorticoid response in a mutant human androgen receptor (ARccr) derived from an androgen-independent prostate cancer | | Descriptor: | 9ALPHA-FLUOROCORTISOL, ANDROGEN RECEPTOR, PHOSPHATE ION | | Authors: | Matias, P.M, Carrondo, M.A, Coelho, R, Thomaz, M, Zhao, X.-Y, Wegg, A, Crusius, K, Egner, U, Donner, P. | | Deposit date: | 2001-12-27 | | Release date: | 2003-01-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Basis for the Glucocorticoid Response in a Mutant Human Androgen Receptor (Ar(Ccr)) Derived from an Androgen-Independent Prostate Cancer
J.Med.Chem., 45, 2002
|
|
1GS5
 
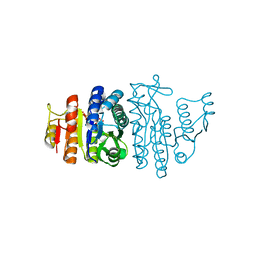 | | N-acetyl-L-glutamate kinase from Escherichia coli complexed with its substrate N-acetylglutamate and its substrate analog AMPPNP | | Descriptor: | ACETYLGLUTAMATE KINASE, MAGNESIUM ION, N-ACETYL-L-GLUTAMATE, ... | | Authors: | Ramon-Maiques, S, Marina, A, Gil-Ortiz, F, Fita, I, Rubio, V. | | Deposit date: | 2001-12-28 | | Release date: | 2002-05-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of Acetylglutamate Kinase, a Key Enzyme for Arginine Biosynthesis and a Prototype for the Amino Acid Kinase Enzyme Family, During Catalysis
Structure, 10, 2002
|
|
1GS6
 
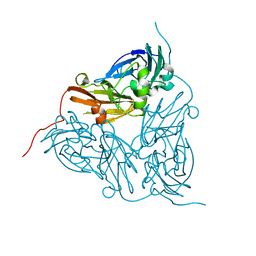 | | Crystal structure of M144A mutant of Alcaligenes xylosoxidans Nitrite Reductase | | Descriptor: | COPPER (II) ION, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE, MAGNESIUM ION | | Authors: | Ellis, M.J, Prudencio, M, Dodd, F.E, Strange, R.W, Sawers, G, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2002-01-02 | | Release date: | 2002-02-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biochemical and Crystallographic Studies of the met144Ala, Asp92Asn and His254Phe Mutants of the Nitrite Reductase from Alcaligenes Xylosoxidans Provide Insight Into the Enzyme Mechanism.
J.Mol.Biol., 316, 2002
|
|
1GS7
 
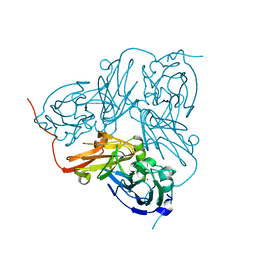 | | Crystal structure of H254F mutant of Alcaligenes xylosoxidans Nitrite Reductase | | Descriptor: | COPPER (II) ION, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE, ZINC ION | | Authors: | Ellis, M.J, Prudencio, M, Dodd, F.E, Strange, R.W, Sawers, G, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2002-01-02 | | Release date: | 2002-02-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Biochemical and Crystallographic Studies of the met144Ala, Asp92Asn and His254Phe Mutants of the Nitrite Reductase from Alcaligenes Xylosoxidans Provide Insight Into the Enzyme Mechanism.
J.Mol.Biol., 316, 2002
|
|
