1D29
 
 | |
1D36
 
 | | FACILE FORMATION OF A CROSSLINKED ADDUCT BETWEEN DNA AND THE DAUNORUBICIN DERIVATIVE MAR70 MEDIATED BY FORMALDEHYDE: MOLECULAR STRUCTURE OF THE MAR70-D(CGTNACG) COVALENT ADDUC | | Descriptor: | 4'-EPI-4'-(2-DEOXYFUCOSE)DAUNOMYCIN, DNA (5'-D(*CP*GP*TP*AP*CP*G)-3'), MAGNESIUM ION | | Authors: | Gao, Y.-G, Liaw, Y.-C, Li, Y.-K, Van Der Marel, G.A, Van Boom, J.H, Wang, A.H.-J. | | Deposit date: | 1991-04-23 | | Release date: | 1992-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Facile formation of a crosslinked adduct between DNA and the daunorubicin derivative MAR70 mediated by formaldehyde: molecular structure of the MAR70-d(CGTnACG) covalent adduct.
Proc.Natl.Acad.Sci.USA, 88, 1991
|
|
1D46
 
 | | DNA DODECAMER C-G-C-G-A-A-T-T-C-G-C-G/HOECHST 33258 COMPLEX:-100 DEGREES C, PIPERAZINE DOWN | | Descriptor: | 2'-(4-HYDROXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Quintana, J.R, Lipanov, A.A, Dickerson, R.E. | | Deposit date: | 1991-06-04 | | Release date: | 1992-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Low-temperature crystallographic analyses of the binding of Hoechst 33258 to the double-helical DNA dodecamer C-G-C-G-A-A-T-T-C-G-C-G.
Biochemistry, 30, 1991
|
|
6ENL
 
 | |
3LHM
 
 | |
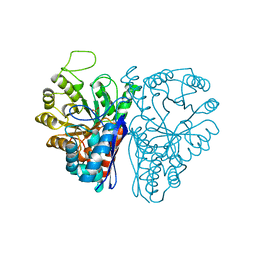
4ENL
 
 | |
3FBP
 
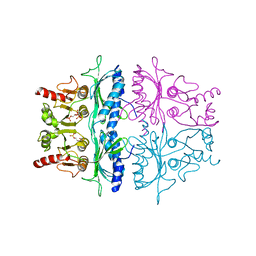 | | STRUCTURE REFINEMENT OF FRUCTOSE-1,6-BISPHOSPHATASE AND ITS FRUCTOSE 2,6-BISPHOSPHATE COMPLEX AT 2.8 ANGSTROMS RESOLUTION | | Descriptor: | 2,6-di-O-phosphono-beta-D-fructofuranose, FRUCTOSE 1,6-BISPHOSPHATASE | | Authors: | Ke, H, Thorpe, C.M, Seaton, B.A, Marcus, F, Lipscomb, W.N. | | Deposit date: | 1990-06-07 | | Release date: | 1992-04-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure refinement of fructose-1,6-bisphosphatase and its fructose 2,6-bisphosphate complex at 2.8 A resolution.
J.Mol.Biol., 212, 1990
|
|
2D34
 
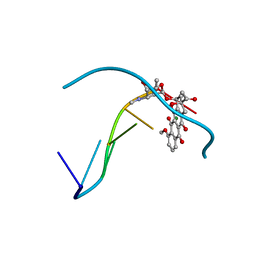 | | FORMALDEHYDE CROSS-LINKS DAUNORUBICIN AND DNA EFFICIENTLY: HPLC AND X-RAY DIFFRACTION STUDIES | | Descriptor: | 5'-D(*CP*GP*TP*(A35)P*CP*G)-3', DAUNOMYCIN, MAGNESIUM ION | | Authors: | Wang, A.H.-J, Gao, Y.-G, Liaw, Y.-C, Li, Y.-K. | | Deposit date: | 1991-05-23 | | Release date: | 1992-04-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Formaldehyde cross-links daunorubicin and DNA efficiently: HPLC and X-ray diffraction studies.
Biochemistry, 30, 1991
|
|
2FBP
 
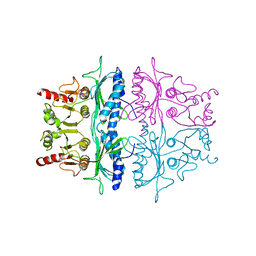 | | STRUCTURE REFINEMENT OF FRUCTOSE-1,6-BISPHOSPHATASE AND ITS FRUCTOSE 2,6-BISPHOSPHATE COMPLEX AT 2.8 ANGSTROMS RESOLUTION | | Descriptor: | FRUCTOSE 1,6-BISPHOSPHATASE | | Authors: | Ke, H, Thorpe, C.M, Seaton, B.A, Marcus, F, Lipscomb, W.N. | | Deposit date: | 1990-06-07 | | Release date: | 1992-04-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure refinement of fructose-1,6-bisphosphatase and its fructose 2,6-bisphosphate complex at 2.8 A resolution.
J.Mol.Biol., 212, 1990
|
|
1D43
 
 | | DNA DODECAMER C-G-C-G-A-A-T-T-C-G-C-G/HOECHST 33258 COMPLEX: 0 DEGREES C, PIPERAZINE UP | | Descriptor: | 2'-(4-HYDROXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Quintana, J.R, Lipanov, A.A, Dickerson, R.E. | | Deposit date: | 1991-06-04 | | Release date: | 1992-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Low-temperature crystallographic analyses of the binding of Hoechst 33258 to the double-helical DNA dodecamer C-G-C-G-A-A-T-T-C-G-C-G.
Biochemistry, 30, 1991
|
|
1D49
 
 | | THE STRUCTURE OF A B-DNA DECAMER WITH A CENTRAL T-A STEP: C-G-A-T-T-A-A-T-C-G | | Descriptor: | DNA (5'-D(*CP*GP*AP*TP*TP*AP*AP*TP*CP*G)-3'), MAGNESIUM ION | | Authors: | Quintana, J.R, Grzeskowiak, K, Yanagi, K, Dickerson, R.E. | | Deposit date: | 1991-09-17 | | Release date: | 1992-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of a B-DNA decamer with a central T-A step: C-G-A-T-T-A-A-T-C-G.
J.Mol.Biol., 225, 1992
|
|
21BI
 
 | |
1D30
 
 | | THE STRUCTURE OF DAPI BOUND TO DNA | | Descriptor: | 6-AMIDINE-2-(4-AMIDINO-PHENYL)INDOLE, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Larsen, T, Goodsell, D.S, Cascio, D, Grzeskowiak, K, Dickerson, R.E. | | Deposit date: | 1991-01-04 | | Release date: | 1992-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structure of DAPI bound to DNA.
J.Biomol.Struct.Dyn., 7, 1989
|
|
1D41
 
 | |
1D48
 
 | | STRUCTURE OF THE PURE-SPERMINE FORM OF Z-DNA (MAGNESIUM FREE) AT 1 ANGSTROM RESOLUTION | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*CP*G)-3'), SPERMINE | | Authors: | Egli, M, Williams, L.D, Gao, Q, Rich, A. | | Deposit date: | 1991-09-11 | | Release date: | 1992-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structure of the pure-spermine form of Z-DNA (magnesium free) at 1-A resolution.
Biochemistry, 30, 1991
|
|
2LHM
 
 | |
1NDN
 
 | | MOLECULAR STRUCTURE OF NICKED DNA. MODEL T4 | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*AP*AP*CP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*GP*TP*T)-3'), DNA (5'-D(*TP*TP*CP*GP*CP*G)-3') | | Authors: | Aymani, J, Coll, M, Van Der Marel, G.A, Van Boom, J.H, Wang, A.H.-J, Rich, A. | | Deposit date: | 1992-01-15 | | Release date: | 1992-07-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular structure of nicked DNA: a substrate for DNA repair enzymes.
Proc.Natl.Acad.Sci.USA, 87, 1990
|
|
9CA2
 
 | |
2YCC
 
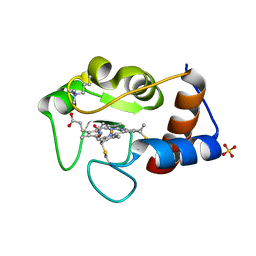 | |
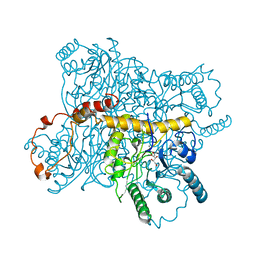
4XIS
 
 | |
3NN9
 
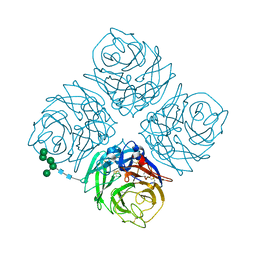 | | REFINED ATOMIC STRUCTURES OF N9 SUBTYPE INFLUENZA VIRUS NEURAMINIDASE AND ESCAPE MUTANTS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, NEURAMINIDASE N9, ... | | Authors: | Tulip, W.R, Varghese, J.N, Baker, A.T, Vandonkelaar, A, Laver, W.G, Webster, R.G, Colman, P.M. | | Deposit date: | 1991-03-28 | | Release date: | 1992-07-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Refined atomic structures of N9 subtype influenza virus neuraminidase and escape mutants.
J.Mol.Biol., 221, 1991
|
|
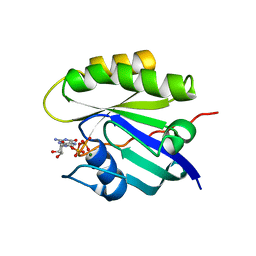
4Q21
 
 | |
1RMS
 
 | | CRYSTAL STRUCTURES OF RIBONUCLEASE MS COMPLEXED WITH 3'-GUANYLIC ACID A GP*C ANALOGUE, 2'-DEOXY-2'-FLUOROGUANYLYL-3',5'-CYTIDINE | | Descriptor: | GUANOSINE-3'-MONOPHOSPHATE, RIBONUCLEASE MS | | Authors: | Nonaka, T, Mitsui, Y, Nakamura, K.T. | | Deposit date: | 1991-12-02 | | Release date: | 1992-07-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of ribonuclease Ms (as a ribonuclease T1 homologue) complexed with a guanylyl-3',5'-cytidine analogue.
Biochemistry, 32, 1993
|
|
6LZM
 
 | | COMPARISON OF THE CRYSTAL STRUCTURE OF BACTERIOPHAGE T4 LYSOZYME AT LOW, MEDIUM, AND HIGH IONIC STRENGTHS | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Bell, J.A, Wilson, K, Zhang, X.-J, Faber, H.R, Nicholson, H, Matthews, B.W. | | Deposit date: | 1991-01-25 | | Release date: | 1992-07-15 | | Last modified: | 2021-06-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Comparison of the crystal structure of bacteriophage T4 lysozyme at low, medium, and high ionic strengths.
Proteins, 10, 1991
|
|
2HIP
 
 | | THE MOLECULAR STRUCTURE OF THE HIGH POTENTIAL IRON-SULFUR PROTEIN ISOLATED FROM ECTOTHIORHODOSPIRA HALOPHILA DETERMINED AT 2.5-ANGSTROMS RESOLUTION | | Descriptor: | HIGH POTENTIAL IRON SULFUR PROTEIN, IRON/SULFUR CLUSTER | | Authors: | Breiter, D.R, Meyer, T.E, Rayment, I, Holden, H.M. | | Deposit date: | 1991-06-24 | | Release date: | 1992-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The molecular structure of the high potential iron-sulfur protein isolated from Ectothiorhodospira halophila determined at 2.5-A resolution.
J.Biol.Chem., 266, 1991
|
|
