1R8J
 
 | |
1R9F
 
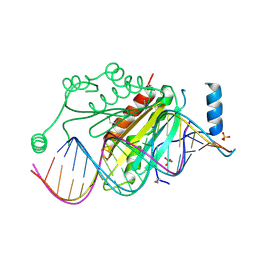 | | Crystal structure of p19 complexed with 19-bp small interfering RNA | | Descriptor: | 5'-R(*CP*GP*UP*AP*CP*GP*CP*GP*GP*AP*AP*UP*AP*CP*UP*UP*CP*GP*AP*UP*U)-3', 5'-R(*UP*CP*GP*AP*AP*GP*UP*AP*UP*UP*CP*CP*GP*CP*GP*UP*AP*CP*GP*UP*U)-3', Core protein P19, ... | | Authors: | Ye, K, Malinina, L, Patel, D.J. | | Deposit date: | 2003-10-28 | | Release date: | 2004-01-27 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Recognition of small interfering RNA by a viral suppressor of RNA
Nature, 426, 2003
|
|
7P7I
 
 | | Native structure of N-acetylglucosamine kinase from Plesiomonas shigelloides | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Roy, S, Isupov, M.N, Harmer, N.J, Ames, J.R. | | Deposit date: | 2021-07-19 | | Release date: | 2022-07-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Spinning sugars in antigen biosynthesis: characterization of the Coxiella burnetii and Streptomyces griseus TDP-sugar epimerases
J.Biol.Chem., 2022
|
|
1RH3
 
 | |
7P55
 
 | |
1RE9
 
 | | CRYSTAL STRUCTURE OF CYTOCHROME P450-CAM WITH A FLUORESCENT PROBE D-8-AD (ADAMANTANE-1-CARBOXYLIC ACID-5-DIMETHYLAMINO-NAPHTHALENE-1-SULFONYLAMINO-OCTYL-AMIDE) | | Descriptor: | 1,2-ETHANEDIOL, ADAMANTANE-1-CARBOXYLIC ACID-5-DIMETHYLAMINO-NAPHTHALENE-1-SULFONYLAMINO-OCTYL-AMIDE, Cytochrome P450-cam, ... | | Authors: | Hays, A.-M.A, Dunn, A.R, Gray, H.B, Stout, C.D, Goodin, D.B. | | Deposit date: | 2003-11-06 | | Release date: | 2004-11-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Conformational states of cytochrome P450cam revealed by trapping of synthetic molecular wires.
J.Mol.Biol., 344, 2004
|
|
7PB3
 
 | | Structural and Functional analysis of the Proline Racemase (ProR) from the Gram-positive bacterium Acetoanaerobium sticklandii | | Descriptor: | PYRROLE-2-CARBOXYLATE, Proline racemase A (AsProR) | | Authors: | Najmudin, S, Pan, X.-S, McAuley, K.E, Fisher, L.M, Sanderson, M.R. | | Deposit date: | 2021-07-30 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.841 Å) | | Cite: | Structural and Functional analysis of the Proline Racemase (ProR) from the Gram-positive bacterium Acetoanaerobium sticklandii
To Be Published
|
|
7PA2
 
 | | PARK7 with inhibitor 8RK64 | | Descriptor: | (3~{S})-~{N}-[5-[2-[(azanylidene-$l^{4}-azanylidene)amino]ethanoyl]-6,7-dihydro-4~{H}-[1,3]thiazolo[5,4-c]pyridin-2-yl]-1-(iminomethyl)pyrrolidine-3-carboxamide, Parkinson disease protein 7 | | Authors: | Kim, R.Q, Jia, Y, Sapmaz, A, Geurink, P.P. | | Deposit date: | 2021-07-28 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Chemical Toolkit for PARK7: Potent, Selective, and High-Throughput.
J.Med.Chem., 65, 2022
|
|
1RF7
 
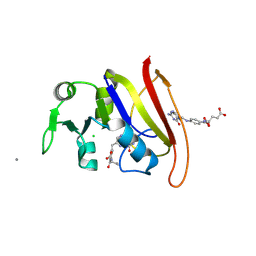 | | STRUCTURE OF DIHYDROFOLATE REDUCTASE COMPLEXED WITH DIHYDROFOLATE | | Descriptor: | CHLORIDE ION, DIHYDROFOLATE REDUCTASE, DIHYDROFOLIC ACID, ... | | Authors: | Sawaya, M.R. | | Deposit date: | 1996-10-24 | | Release date: | 1997-03-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Loop and subdomain movements in the mechanism of Escherichia coli dihydrofolate reductase: crystallographic evidence.
Biochemistry, 36, 1997
|
|
1RKJ
 
 | | Solution structure of the complex formed by the two N-terminal RNA-binding domains of nucleolin and a pre-rRNA target | | Descriptor: | 5'-R(*GP*GP*AP*UP*GP*CP*CP*UP*CP*CP*CP*GP*AP*GP*UP*GP*CP*AP*UP*CP*C)-3', Nucleolin | | Authors: | Johansson, C, Finger, L.D, Trantirek, L, Mueller, T.D, Kim, S, Laird-Offringa, I.A, Feigon, J. | | Deposit date: | 2003-11-21 | | Release date: | 2004-04-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the complex formed by the two N-terminal RNA-binding domains of nucleolin and a pre-rRNA target.
J.Mol.Biol., 337, 2004
|
|
7P9W
 
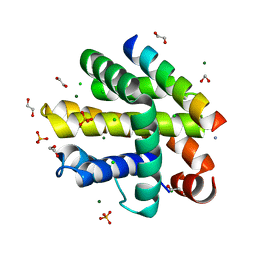 | | Epstein-Barr virus encoded apoptosis regulator BHRF1 in complex with Puma BH3 | | Descriptor: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, AMMONIUM ION, ... | | Authors: | Suraweera, C.D, Hinds, M.G, Kvansakul, M. | | Deposit date: | 2021-07-28 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.00010061 Å) | | Cite: | Crystal Structures of Epstein-Barr Virus Bcl-2 Homolog BHRF1 Bound to Bid and Puma BH3 Motif Peptides.
Viruses, 14, 2022
|
|
7PA3
 
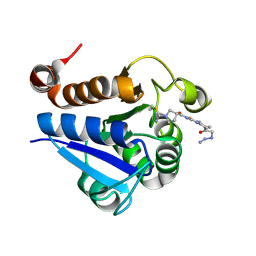 | | PARK7 with covalent inhibitor JYQ-88 | | Descriptor: | (3~{S})-~{N}-[5-[2-[(azanylidene-$l^{4}-azanylidene)amino]ethanoyl]-6,7-dihydro-4~{H}-[1,3]thiazolo[5,4-c]pyridin-2-yl]-1-(iminomethyl)pyrrolidine-3-carboxamide, Parkinson disease protein 7 | | Authors: | Kim, R.Q, Jia, Y, Sapmaz, A, Geurink, P.P. | | Deposit date: | 2021-07-28 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Chemical Toolkit for PARK7: Potent, Selective, and High-Throughput.
J.Med.Chem., 65, 2022
|
|
7PA0
 
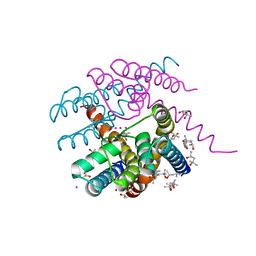 | | NaK C-DI F92A mutant with Rb+ and K+ | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, POTASSIUM ION, Potassium channel protein, ... | | Authors: | Minniberger, S, Plested, A.J.R. | | Deposit date: | 2021-07-28 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Asymmetry and Ion Selectivity Properties of Bacterial Channel NaK Mutants Derived from Ionotropic Glutamate Receptors.
J.Mol.Biol., 435, 2023
|
|
7PA1
 
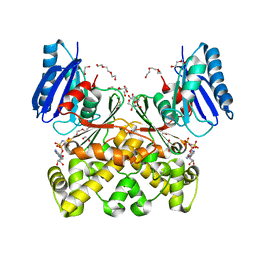 | | Structure of N-acetylglucosamine kinase from Plesiomonas shigelloides in complex with AMP-PNP in the absence of N-acetylglucoseamine substrate | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Roy, S, Isupov, M.N, Harmer, N.J, Ames, J.R. | | Deposit date: | 2021-07-28 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Spinning sugars in antigen biosynthesis: characterization of the Coxiella burnetii and Streptomyces griseus TDP-sugar epimerases
J.Biol.Chem., 2022
|
|
2B49
 
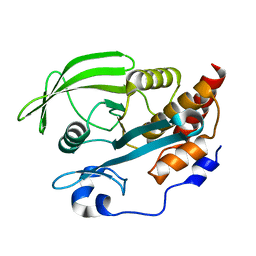 | | Crystal Structure of the Catalytic Domain of Protein Tyrosine Phosphatase, non-receptor Type 3 | | Descriptor: | protein tyrosine phosphatase, non-receptor type 3 | | Authors: | Ugochukwu, E, Arrowsmith, C, Barr, A, Bunkoczi, G, Das, S, Debreczeni, J, Edwards, A, Eswaran, J, Knapp, S, Sundstrom, M, Turnbull, A, von Delft, F, Weigelt, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-09-23 | | Release date: | 2005-10-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Large-scale structural analysis of the classical human protein tyrosine phosphatome.
Cell(Cambridge,Mass.), 136, 2009
|
|
7PB6
 
 | |
1RG7
 
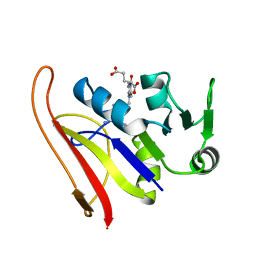 | | DIHYDROFOLATE REDUCTASE COMPLEXED WITH METHOTREXATE | | Descriptor: | DIHYDROFOLATE REDUCTASE, METHOTREXATE | | Authors: | Sawaya, M.R, Kraut, J. | | Deposit date: | 1996-11-27 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Loop and subdomain movements in the mechanism of Escherichia coli dihydrofolate reductase: crystallographic evidence.
Biochemistry, 36, 1997
|
|
1RN7
 
 | | Structure of human cystatin D | | Descriptor: | Cystatin D | | Authors: | Alvarez-Fernandez, M, Liang, Y.H, Abrahamson, M, Su, X.D. | | Deposit date: | 2003-11-30 | | Release date: | 2004-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of human cystatin D, a cysteine peptidase inhibitor with restricted inhibition profile.
J.Biol.Chem., 280, 2005
|
|
7PB7
 
 | |
7PBH
 
 | |
7PC1
 
 | |
7OQ9
 
 | |
7OQA
 
 | |
1RIK
 
 | | E6-binding zinc finger (E6apc1) | | Descriptor: | E6apc1 peptide | | Authors: | Liu, Y, Liu, Z, Androphy, E, Chen, J, Baleja, J.D. | | Deposit date: | 2003-11-17 | | Release date: | 2004-08-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Design and characterization of helical peptides that inhibit the E6 protein of papillomavirus.
Biochemistry, 43, 2004
|
|
1RU7
 
 | | 1934 Human H1 Hemagglutinin | | Descriptor: | hemagglutinin | | Authors: | Skehel, J.J, Gamblin, S.J, Haire, L.F, Russell, R.J, Stevens, D.J, Xiao, B, Ha, Y, Vasisht, N, Steinhauer, D.A, Daniels, R.S. | | Deposit date: | 2003-12-11 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure and receptor binding properties of the 1918 influenza hemagglutinin.
Science, 303, 2004
|
|
