4NTD
 
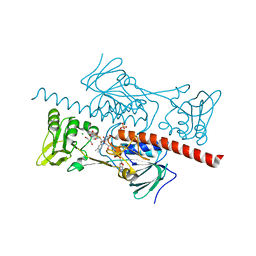 | | Crystal structure of HlmI | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLUTATHIONE, ... | | Authors: | Scharf, D.H, Groll, M, Habel, A, Heinekamp, T, Hertweck, C, Brakhage, A.A, Huber, E.M. | | Deposit date: | 2013-12-02 | | Release date: | 2014-03-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Flavoenzyme-Catalyzed Formation of Disulfide Bonds in Natural Products
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
3UI2
 
 | |
1XU6
 
 | | Structure of the C-terminal domain from Trypanosoma brucei Variant Surface Glycoprotein MITat1.2 | | Descriptor: | Variant surface glycoprotein MITAT 1.2 | | Authors: | Chattopadhyay, A, Jones, N.G, Nietlispach, D, Nielsen, P.R, Voorheis, H.P, Mott, H.R, Carrington, M. | | Deposit date: | 2004-10-25 | | Release date: | 2004-11-30 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structure of the C-terminal domain from Trypanosoma brucei variant surface glycoprotein MITat1.2
J.Biol.Chem., 280, 2004
|
|
4JJ2
 
 | | High resolution structure of a C-terminal fragment of the T4 phage gp5 beta-helix | | Descriptor: | 9-OCTADECENOIC ACID, MAGNESIUM ION, PALMITIC ACID, ... | | Authors: | Buth, S.A, Leiman, P.G, Boudko, S.P. | | Deposit date: | 2013-03-07 | | Release date: | 2014-03-12 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Structure and Biophysical Properties of a Triple-Stranded Beta-Helix Comprising the Central Spike of Bacteriophage T4.
Viruses, 7, 2015
|
|
1ZJR
 
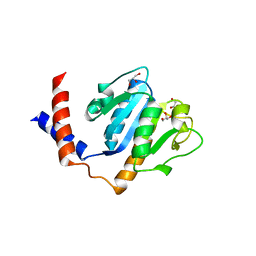 | | Crystal Structure of A. aeolicus TrmH/SpoU tRNA modifying enzyme | | Descriptor: | GLYCEROL, SULFATE ION, tRNA (Guanosine-2'-O-)-methyltransferase | | Authors: | Pleshe, E, Truesdell, J, Batey, R.T. | | Deposit date: | 2005-04-30 | | Release date: | 2005-08-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of a class II TrmH tRNA-modifying enzyme from Aquifex aeolicus.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
2JWG
 
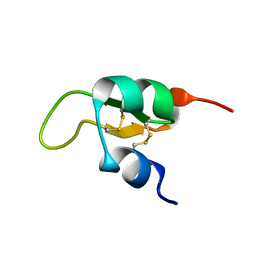 | | Structure of a Glycosylphosphatidylinositol-anchored Domain from a Trypanosome Variant Surface Glycoprotein | | Descriptor: | Variant surface glycoprotein ILTAT 1.24 | | Authors: | Jones, N.G, Nietlispach, D, Sharma, R, Burke, D.F, Eyres, I, Mues, M, Mott, H.R, Carrington, M. | | Deposit date: | 2007-10-12 | | Release date: | 2007-11-13 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structure of a Glycosylphosphatidylinositol-anchored Domain from a Trypanosome Variant Surface Glycoprotein
J.Biol.Chem., 283, 2008
|
|
2I9D
 
 | |
2H1N
 
 | | 3.0 A X-ray structure of putative oligoendopeptidase F: crystals grown by vapor diffusion technique | | Descriptor: | Oligoendopeptidase F, UNKNOWN LIGAND, ZINC ION | | Authors: | Gerdts, C.J, Tereshko, V, Dementieva, I, Collart, F, Joachimiak, A, Kossiakoff, A, Ismagilov, R.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-05-16 | | Release date: | 2006-06-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Time-Controlled Microfluidic Seeding in nL-Volume Droplets To Separate Nucleation and Growth Stages of Protein Crystallization.
Angew.Chem.Int.Ed.Engl., 45, 2006
|
|
2JM4
 
 | |
2JWH
 
 | | Structure of a Glycosylphosphatidylinositol-anchored Domain from a Trypanosome Variant Surface Glycoprotein | | Descriptor: | Variant surface glycoprotein ILTAT 1.24 | | Authors: | Jones, N.G, Nietlispach, D, Sharma, R, Burke, D.F, Eyres, I, Mues, M, Mott, H.R, Carrington, M. | | Deposit date: | 2007-10-12 | | Release date: | 2007-11-13 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structure of a Glycosylphosphatidylinositol-anchored Domain from a Trypanosome Variant Surface Glycoprotein
J.Biol.Chem., 283, 2008
|
|
