2O69
 
 | |
8A82
 
 | | Fe(II)/aKG-dependent halogenase OocPQ | | Descriptor: | Cupin_8 domain-containing protein, FE (III) ION, GLYCEROL, ... | | Authors: | Fraley, A.E, Meoded, R.A, Schmalhofer, M, Bergande, C, Groll, M, Piel, J. | | Deposit date: | 2022-06-21 | | Release date: | 2023-03-08 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Heterocomplex structure of a polyketide synthase component involved in modular backbone halogenation.
Structure, 31, 2023
|
|
3RTT
 
 | | Human MMP-12 catalytic domain in complex with*(R)-N*-Hydroxy-1-(phenethylsulfonyl)pyrrolidine-2-carboxamide | | Descriptor: | CALCIUM ION, Macrophage metalloelastase, N-hydroxy-1-[(2-phenylethyl)sulfonyl]-D-prolinamide, ... | | Authors: | Bertini, I, Calderone, V, Fragai, M, Luchinat, C, Mori, M, Nativi, C. | | Deposit date: | 2011-05-04 | | Release date: | 2012-07-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Contribution of ligand free energy of solvation to design new potent MMPs inhibitors.
J.Med.Chem., 2012
|
|
2UUB
 
 | | Structure of the Thermus thermophilus 30S ribosomal subunit complexed with a Valine-ASL with cmo5U in position 34 bound to an mRNA with a GUU-codon in the A-site and paromomycin. | | Descriptor: | 16S Ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Weixlbaumer, A, Murphy, F.V, Dziergowska, A, Malkiewicz, A, Vendeix, F.A.P, Agris, P.F, Ramakrishnan, V. | | Deposit date: | 2007-03-01 | | Release date: | 2007-05-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanism for Expanding the Decoding Capacity of Transfer Rnas by Modification of Uridines
Nat.Struct.Mol.Biol., 14, 2007
|
|
2UUA
 
 | | Structure of the Thermus thermophilus 30S ribosomal subunit complexed with a Valine-ASL with cmo5U in position 34 bound to an mRNA with a GUC-codon in the A-site and paromomycin. | | Descriptor: | 16S RRNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Weixlbaumer, A, Murphy, F.V, Dziergowska, A, Malkiewicz, A, Vendeix, F.A.P, Agris, P.F, Ramakrishnan, V. | | Deposit date: | 2007-03-01 | | Release date: | 2007-05-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mechanism for expanding the decoding capacity of transfer RNAs by modification of uridines.
Nat. Struct. Mol. Biol., 14, 2007
|
|
5UKQ
 
 | | Structure of unliganded anti-gp120 CD4bs antibody DH522.2 Fab | | Descriptor: | DH522.2 Fab fragment heavy chain, DH522.2 Fab fragment light chain, GLYCEROL | | Authors: | Nicely, N.I. | | Deposit date: | 2017-01-23 | | Release date: | 2017-12-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Initiation of HIV neutralizing B cell lineages with sequential envelope immunizations.
Nat Commun, 8, 2017
|
|
1XRR
 
 | | Crystal structure of active site F1-mutant E245Q soaked with peptide Pro-Pro | | Descriptor: | PROLINE, Proline iminopeptidase | | Authors: | Goettig, P, Brandstetter, H, Groll, M, Goehring, W, Konarev, P.V, Svergun, D.I, Huber, R, Kim, J.-S. | | Deposit date: | 2004-10-15 | | Release date: | 2005-07-12 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray snapshots of peptide processing in mutants of tricorn-interacting factor F1 from Thermoplasma acidophilum
J.Biol.Chem., 280, 2005
|
|
1PVO
 
 | |
3J15
 
 | | Model of ribosome-bound archaeal Pelota and ABCE1 | | Descriptor: | ABC transporter ATP-binding protein, ADENOSINE-5'-DIPHOSPHATE, IRON/SULFUR CLUSTER, ... | | Authors: | Becker, T, Franckenberg, S, Wickles, S, Shoemaker, C.J, Anger, A.M, Armache, J.-P, Sieber, H, Ungewickell, C, Berninghausen, O, Daberkow, I, Karcher, A, Thomm, M, Hopfner, K.-P, Green, R, Beckmann, R. | | Deposit date: | 2011-12-12 | | Release date: | 2012-02-22 | | Last modified: | 2018-08-22 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Structural basis of highly conserved ribosome recycling in eukaryotes and archaea.
Nature, 482, 2012
|
|
6ZLU
 
 | |
5XD9
 
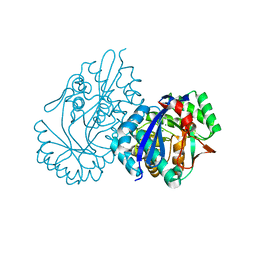 | | Crystal structure analysis of 3,6-anhydro-L-galactonate cycloisomerase | | Descriptor: | 3,6-anhydro-alpha-L-galactonate cycloisomerase, MAGNESIUM ION | | Authors: | Lee, S, Choi, I.-G, Kim, H.-Y. | | Deposit date: | 2017-03-27 | | Release date: | 2017-09-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure analysis of 3,6-anhydro-l-galactonate cycloisomerase suggests emergence of novel substrate specificity in the enolase superfamily
Biochem. Biophys. Res. Commun., 491, 2017
|
|
1XPO
 
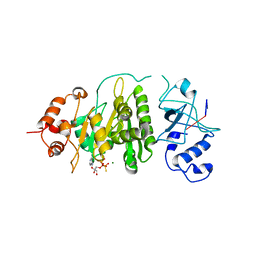 | | Structural mechanism of inhibition of the Rho transcription termination factor by the antibiotic bicyclomycin | | Descriptor: | 5'-R(*CP*UP*CP*UP*CP*UP*CP*U)-3', BICYCLOMYCIN, MAGNESIUM ION, ... | | Authors: | Skordalakes, E, Brogan, A.P, Park, B.S, Kohn, H, Berger, J.M. | | Deposit date: | 2004-10-09 | | Release date: | 2005-02-08 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural mechanism of inhibition of the rho transcription termination factor by the antibiotic bicyclomycin
Structure, 13, 2005
|
|
6ZS3
 
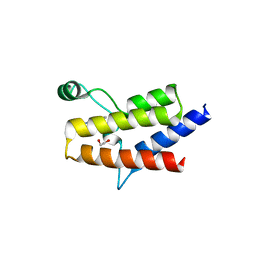 | | Crystal structure of the fifth bromodomain of human protein polybromo-1 in complex with 2-(6-amino-5-(piperazin-1-yl)pyridazin-3-yl)phenol | | Descriptor: | 1,2-ETHANEDIOL, 2-(6-azanyl-5-piperazin-4-ium-1-yl-pyridazin-3-yl)phenol, Protein polybromo-1 | | Authors: | Preuss, F, Joerger, A.C, Wanior, M, Kraemer, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-07-15 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Pan-SMARCA/PB1 Bromodomain Inhibitors and Their Role in Regulating Adipogenesis.
J.Med.Chem., 63, 2020
|
|
1PX7
 
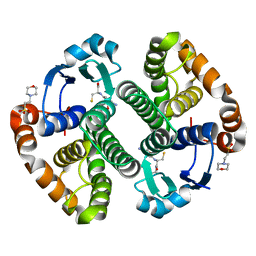 | | A folding mutant of human class pi glutathione transferase, created by mutating aspartate 153 of the wild-type protein to glutamate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLUTATHIONE, ... | | Authors: | Kong, G.K.-W, Polekhina, G, McKinstry, W.J, Parker, M.W, Dragani, B, Aceto, A, Paludi, D, Principe, D.R, Mannervik, B, Stenberg, G. | | Deposit date: | 2003-07-02 | | Release date: | 2003-07-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | The multi-functional role of a highly conserved aspartic acid residue in glutathione transferase P1-1
To be Published
|
|
1XKU
 
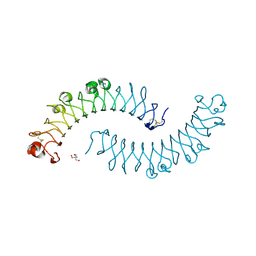 | | Crystal structure of the dimeric protein core of decorin, the archetypal small leucine-rich repeat proteoglycan | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Decorin | | Authors: | Scott, P.G, McEwan, P.A, Dodd, C.M, Bergmann, E.M, Bishop, P.N, Bella, J. | | Deposit date: | 2004-09-29 | | Release date: | 2004-11-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of the dimeric protein core of decorin, the archetypal small leucine-rich repeat proteoglycan
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
6ZM1
 
 | |
1Q57
 
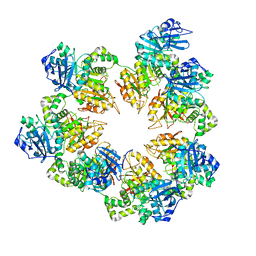 | | The Crystal Structure of the Bifunctional Primase-Helicase of Bacteriophage T7 | | Descriptor: | DNA primase/helicase | | Authors: | Toth, E.A, Li, Y, Sawaya, M.R, Cheng, Y, Ellenberger, T. | | Deposit date: | 2003-08-06 | | Release date: | 2003-11-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | The Crystal Structure of the Bifunctional Primase-Helicase of Bacteriophage T7
Mol.Cell, 12, 2003
|
|
1XRQ
 
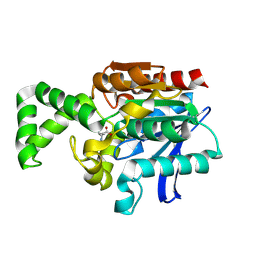 | | Crystal structure of active site F1-mutant E245Q soaked with peptide Phe-Leu | | Descriptor: | LEUCINE, Proline iminopeptidase | | Authors: | Goettig, P, Brandstetter, H, Groll, M, Goehring, W, Konarev, P.V, Svergun, D.I, Huber, R, Kim, J.-S. | | Deposit date: | 2004-10-15 | | Release date: | 2005-07-12 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray snapshots of peptide processing in mutants of tricorn-interacting factor F1 from Thermoplasma acidophilum
J.Biol.Chem., 280, 2005
|
|
1XQW
 
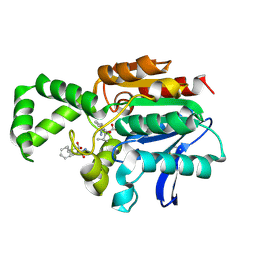 | | Crystal structure of F1-mutant S105A complex with PHE-LEU | | Descriptor: | LEUCINE, PHENYLALANINE, Proline iminopeptidase | | Authors: | Goettig, P, Brandstetter, H, Groll, M, Goehring, W, Konarev, P.V, Svergun, D.I, Huber, R, Kim, J.-S. | | Deposit date: | 2004-10-13 | | Release date: | 2005-07-12 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray snapshots of peptide processing in mutants of tricorn-interacting factor F1 from Thermoplasma acidophilum
J.Biol.Chem., 280, 2005
|
|
1PG1
 
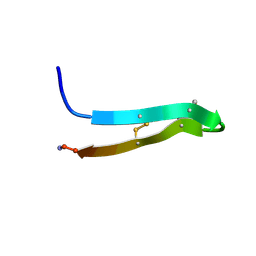 | | PROTEGRIN 1 (PG1) FROM PORCINE LEUKOCYTES, NMR, 20 STRUCTURES | | Descriptor: | PROTEGRIN-1 | | Authors: | Fahrner, R.L, Dieckmann, T, Harwig, S.S.L, Lehrer, R.I, Eisenberg, D, Feigon, J. | | Deposit date: | 1998-03-20 | | Release date: | 1998-05-27 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of protegrin-1, a broad-spectrum antimicrobial peptide from porcine leukocytes.
Chem.Biol., 3, 1996
|
|
6ZG0
 
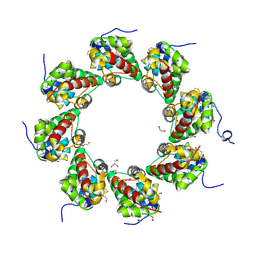 | | SARM1 SAM1-2 domains | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sporny, M, Guez-Haddad, J, Khazma, T, Yaron, A, Dessau, M, Mim, C, Isupov, M.N, Zalk, R, Hons, M, Opatowsky, Y. | | Deposit date: | 2020-06-18 | | Release date: | 2020-11-11 | | Last modified: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Structural basis for SARM1 inhibition and activation under energetic stress.
Elife, 9, 2020
|
|
6ZJK
 
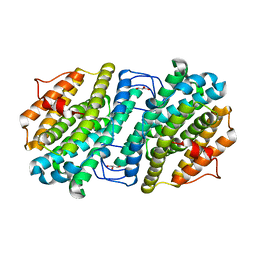 | |
5ZKL
 
 | | Crystal structure of Streptococcus pneumoniae SP_0782 (residues 7-79) in complex with single-stranded DNA dT12 | | Descriptor: | DNA (5'-D(*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), SP_0782 | | Authors: | Lu, G, Li, S, Zhu, J, Yang, Y, Gong, P. | | Deposit date: | 2018-03-24 | | Release date: | 2019-03-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Structural insight into the length-dependent binding of ssDNA by SP_0782 from Streptococcus pneumoniae, reveals a divergence in the DNA-binding interface of PC4-like proteins.
Nucleic Acids Res., 48, 2020
|
|
1XRO
 
 | | Crystal structure of active site F1-mutant E213Q soaked with peptide Phe-Leu | | Descriptor: | LEUCINE, Proline iminopeptidase | | Authors: | Goettig, P, Brandstetter, H, Groll, M, Goehring, W, Konarev, P.V, Svergun, D.I, Huber, R, Kim, J.-S. | | Deposit date: | 2004-10-15 | | Release date: | 2005-07-12 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray snapshots of peptide processing in mutants of tricorn-interacting factor F1 from Thermoplasma acidophilum
J.Biol.Chem., 280, 2005
|
|
2VPQ
 
 | |
