3OMJ
 
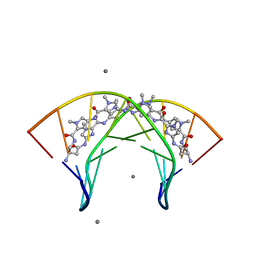 | | Structural Basis for Cyclic Py-Im Polyamide Allosteric Inhibition of Nuclear Receptor Binding | | Descriptor: | (23R,52R)-23,52-diamino-5,11,17,28,34,40,46,57-octamethyl-2,5,8,11,14,17,20,25,28,31,34,37,40,43,46,49,54,57,60,64-icosaazanonacyclo[54.2.1.1~4,7~.1~10,13~.1~16,19~.1~27,30~.1~33,36~.1~39,42~.1~45,48~]hexahexaconta-1(58),4(66),6,10(65),12,16(64),18,27(63),29,33(62),35,39(61),41,45(60),47,56(59)-hexadecaene-3,9,15,21,26,32,38,44,50,55-decone, 5'-D(*CP*(C38)P*AP*GP*TP*AP*CP*TP*GP*G)-3', CALCIUM ION | | Authors: | Chenoweth, D.M. | | Deposit date: | 2010-08-27 | | Release date: | 2010-09-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Structural basis for cyclic py-im polyamide allosteric inhibition of nuclear receptor binding.
J.Am.Chem.Soc., 132, 2010
|
|
4QIO
 
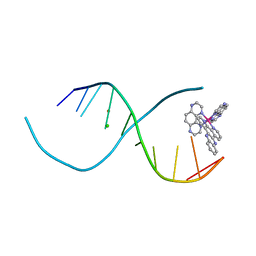 | | Lambda-[Ru(TAP)2(dppz)]2+ bound to d(TCGGCGCCIA) at high resolution | | Descriptor: | 5'-D(*TP*CP*GP*GP*CP*GP*CP*CP*IP*A)-3', BARIUM ION, CHLORIDE ION, ... | | Authors: | Gurung, S.P, Hall, J.P, Cardin, C.J. | | Deposit date: | 2014-06-01 | | Release date: | 2015-06-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Inosine Can Increase DNA's Susceptibility to Photo-oxidation by a Ru II Complex due to Structural Change in the Minor Groove.
Chemistry, 23, 2017
|
|
4F19
 
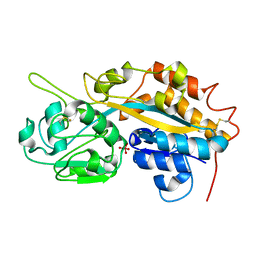 | | Subatomic resolution structure of a high affinity periplasmic phosphate-binding protein (PfluDING) bound with arsenate at pH 4.5 | | Descriptor: | Putative alkaline phosphatase, hydrogen arsenate | | Authors: | Elias, M, Wellner, A, Goldin, K, Chabriere, E, Tawfik, D.S. | | Deposit date: | 2012-05-06 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | The molecular basis of phosphate discrimination in arsenate-rich environments.
Nature, 491, 2012
|
|
1A7Z
 
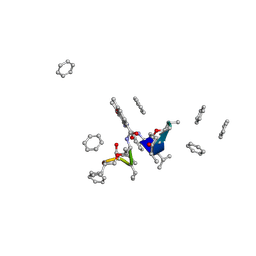 | | CRYSTAL STRUCTURE OF ACTINOMYCIN Z3 | | Descriptor: | ACTINOMYCIN Z3, BENZENE | | Authors: | Schafer, M. | | Deposit date: | 1998-03-19 | | Release date: | 1999-03-23 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Crystal Structures of Actinomycin D and Actinomycin Z3.
Angew.Chem.Int.Ed.Engl., 37, 1998
|
|
3VLA
 
 | | Crystal structure of edgp | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, EDGP | | Authors: | Yoshizawa, T, Shimizu, T, Hirano, H, Sato, M, Hashimoto, H. | | Deposit date: | 2011-11-30 | | Release date: | 2012-04-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Structural basis for inhibition of xyloglucan-specific endo-beta-1,4-glucanase (XEG) by XEG-protein inhibitor
J.Biol.Chem., 287, 2012
|
|
3WGE
 
 | | Crystal structure of ERp46 Trx2 | | Descriptor: | GLYCEROL, Thioredoxin domain-containing protein 5 | | Authors: | Inaba, K, Suzuki, M, Kojima, R. | | Deposit date: | 2013-08-04 | | Release date: | 2014-06-25 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Radically different thioredoxin domain arrangement of ERp46, an efficient disulfide bond introducer of the mammalian PDI family
Structure, 22, 2014
|
|
6IIP
 
 | | Apo-form structure of the HRP3 PWWP domain | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Hepatoma-derived growth factor-related protein 3, SULFATE ION | | Authors: | Wang, Z, Tian, W. | | Deposit date: | 2018-10-07 | | Release date: | 2019-04-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.951 Å) | | Cite: | Apo-form structure of the HRP3 PWWP domain
Nucleic Acids Res., 2019
|
|
5E7W
 
 | | X-ray Structure of Human Recombinant 2Zn insulin at 0.92 Angstrom | | Descriptor: | ACETATE ION, Insulin, N-PROPANOL, ... | | Authors: | Lisgarten, D.R, Naylor, C.E, Palmer, R.A, Lobley, C.M.C. | | Deposit date: | 2015-10-13 | | Release date: | 2016-10-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (0.9519 Å) | | Cite: | Ultra-high resolution X-ray structures of two forms of human recombinant insulin at 100 K.
Chem Cent J, 11, 2017
|
|
5OUK
 
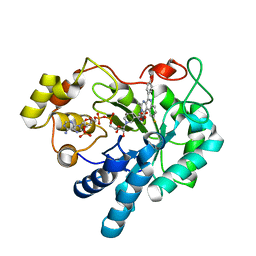 | | Crystal structure of human AKR1B1 complexed with NADP+ and compound 41 | | Descriptor: | 2-[9-fluoranyl-5-(4-methoxyphenyl)-3-methyl-1-oxidanylidene-pyrimido[4,5-c]quinolin-2-yl]ethanoic acid, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Metwally, K, Podjarny, A. | | Deposit date: | 2017-08-24 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (0.959 Å) | | Cite: | Design, synthesis, structure-activity relationships and X-ray structural studies of novel 1-oxopyrimido[4,5-c]quinoline-2-acetic acid derivatives as selective and potent inhibitors of human aldose reductase.
Eur J Med Chem, 152, 2018
|
|
6RKN
 
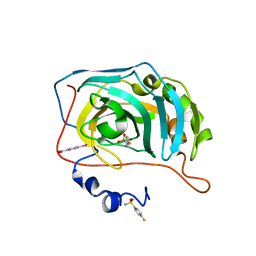 | | Human Carbonic Anhydrase II in complex with a fluorinated Benzenesulfonamide. | | Descriptor: | (4-CARBOXYPHENYL)(CHLORO)MERCURY, 4-FLUOROBENZENESULFONAMIDE, Carbonic anhydrase 2, ... | | Authors: | Gloeckner, S, Heine, A, Klebe, G. | | Deposit date: | 2019-04-30 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.959 Å) | | Cite: | The Influence of Varying Fluorination Patterns on the Thermodynamics and Kinetics of Benzenesulfonamide Binding to Human Carbonic Anhydrase II.
Biomolecules, 10, 2020
|
|
6I74
 
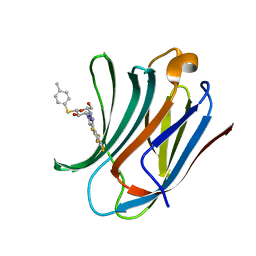 | | Galectin-3C in complex with substituted polyfluoroaryl monothiogalactoside derivative 1 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-(4-methylphenyl)sulfanyl-4-[4-[2,3,4,5,6-pentakis(fluoranyl)phenyl]-1,2,3-triazol-1-yl]oxane-3,5-diol, Galectin-3 | | Authors: | Kumar, R, Peterson, K, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2018-11-15 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.959 Å) | | Cite: | Substituted polyfluoroaryl interactions with an arginine side chain in galectin-3 are governed by steric-, desolvation and electronic conjugation effects.
Org. Biomol. Chem., 17, 2019
|
|
1K5C
 
 | | Endopolygalacturonase I from Stereum purpureum at 0.96 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ENDOPOLYGALACTURONASE, ... | | Authors: | Shimizu, T, Nakatsu, T, Miyairi, K, Okuno, T, Kato, H. | | Deposit date: | 2001-10-10 | | Release date: | 2002-06-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | Active-site architecture of endopolygalacturonase I from Stereum purpureum revealed by crystal structures in native and ligand-bound forms at atomic resolution.
Biochemistry, 41, 2002
|
|
3KFF
 
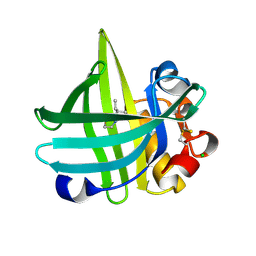 | | Major mouse urinary protein IV complexed with 2-sec-butyl-4,5-dihydrothiazole | | Descriptor: | 2-[(1R)-1-methylpropyl]-4,5-dihydro-1,3-thiazole, 2-[(1S)-1-methylpropyl]-4,5-dihydro-1,3-thiazole, CHLORIDE ION, ... | | Authors: | Perez-Miller, S, Zou, Q, Hurley, T.D. | | Deposit date: | 2009-10-27 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | High resolution X-ray structures of mouse major urinary protein nasal isoform in complex with pheromones.
Protein Sci., 19, 2010
|
|
4R5R
 
 | |
4F18
 
 | | Subatomic resolution structure of a high affinity periplasmic phosphate-binding protein (PfluDING) bound with arsenate at pH 8.5 | | Descriptor: | Putative alkaline phosphatase, hydrogen arsenate | | Authors: | Elias, M, Wellner, A, Goldin, K, Chabriere, E, Tawfik, D.S. | | Deposit date: | 2012-05-06 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | The molecular basis of phosphate discrimination in arsenate-rich environments.
Nature, 491, 2012
|
|
5DP2
 
 | |
3O5Q
 
 | | Fk1 domain mutant A19T of FKBP51, crystal form IV, in presence of DMSO | | Descriptor: | DIMETHYL SULFOXIDE, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Bracher, A, Kozany, C, Thost, A.-K, Hausch, F. | | Deposit date: | 2010-07-28 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | Structural characterization of the PPIase domain of FKBP51, a cochaperone of human Hsp90.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
1U2H
 
 | | X-ray Structure of the N-terminally truncated human APEP-1 | | Descriptor: | Aortic preferentially expressed protein 1 | | Authors: | Manjasetty, B.A, Scheich, C, Roske, Y, Niesen, F.H, Gotz, F, Bussow, K, Heinemann, U. | | Deposit date: | 2004-07-19 | | Release date: | 2005-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | X-ray structure of engineered human Aortic Preferentially Expressed Protein-1 (APEG-1)
Bmc Struct.Biol., 5, 2005
|
|
5ZJC
 
 | |
1UFY
 
 | |
6JPT
 
 | | Crystal structure of human PAC3 homodimer (trigonal form) | | Descriptor: | POTASSIUM ION, Proteasome assembly chaperone 3, THIOCYANATE ION | | Authors: | Satoh, T, Yagi-Utsumi, M, Okamoto, K, Kurimoto, E, Tanaka, K, Kato, K. | | Deposit date: | 2019-03-27 | | Release date: | 2019-05-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | Molecular and Structural Basis of the Proteasome alpha Subunit Assembly Mechanism Mediated by the Proteasome-Assembling Chaperone PAC3-PAC4 Heterodimer.
Int J Mol Sci, 20, 2019
|
|
1LUQ
 
 | |
6J60
 
 | | hnRNP A1 reversible amyloid core GFGGNDNFG (residues 209-217) | | Descriptor: | 9-mer peptide (GFGGNDNFG) from Heterogeneous nuclear ribonucleoprotein A1 | | Authors: | Luo, F, Zhou, H, Gui, X, Li, D, Li, X, Liu, C. | | Deposit date: | 2019-01-12 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON CRYSTALLOGRAPHY (0.96 Å) | | Cite: | Structural basis for reversible amyloids of hnRNPA1 elucidates their role in stress granule assembly.
Nat Commun, 10, 2019
|
|
5ZJ1
 
 | |
8AQP
 
 | |
