9EU6
 
 | | The FK1 domain of FKBP51 in complex with SAFit-analog 23j | | Descriptor: | (1,5-dimethylpyrazol-4-yl)methyl (2~{S})-1-[(2~{S})-2-cyclohexyl-2-(3,4,5-trimethoxyphenyl)ethanoyl]piperidine-2-carboxylate, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Buffa, V, Hausch, F. | | Deposit date: | 2024-03-27 | | Release date: | 2024-06-12 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | 1,4-Pyrazolyl-Containing SAFit-Analogues are Selective FKBP51 Inhibitors With Improved Ligand Efficiency and Drug-Like Profile.
Chemmedchem, 19, 2024
|
|
9EU5
 
 | | SSX structure of Autotaxin at room temperature | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 7alpha-hydroxycholesterol, CALCIUM ION, ... | | Authors: | Eymery, M.C, McCarthy, A.A, Foos, N, Basu, S. | | Deposit date: | 2024-03-27 | | Release date: | 2024-07-31 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | In situ serial crystallography facilitates 96-well plate structural analysis at low symmetry.
Iucrj, 11, 2024
|
|
9ETZ
 
 | | III2IV respiratory supercomplex from Saccharomyces cerevisiae | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSHOCHOLINE, 5-(3,7,11,15,19,23-HEXAMETHYL-TETRACOSA-2,6,10,14,18,22-HEXAENYL)-2,3-DIMETHOXY-6-METHYL-BENZENE-1,4-DIOL, CALCIUM ION, ... | | Authors: | Moe, A, Brzezinski, P. | | Deposit date: | 2024-03-27 | | Release date: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Electron transfer in the respiratory chain at low salinity.
Nat Commun, 15, 2024
|
|
9ETU
 
 | | Archaellum filament from the Halobacterium salinarum deltaAgl27 strain | | Descriptor: | 3-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-beta-D-glucopyranuronic acid-(1-4)-beta-D-glucopyranose, Archaellin | | Authors: | Grossman-Haham, I, Shahar, A. | | Deposit date: | 2024-03-27 | | Release date: | 2024-07-17 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Perturbed N-glycosylation of Halobacterium salinarum archaellum filaments leads to filament bundling and compromised cell motility.
Nat Commun, 15, 2024
|
|
9ETN
 
 | | Crystal structure of murine CRTAC1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Cartilage acidic protein 1, ... | | Authors: | Beugelink, J.W, Hof, H, Janssen, B.J.C. | | Deposit date: | 2024-03-26 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.584 Å) | | Cite: | CRTAC1 has a Compact beta-propeller-TTR Core Stabilized by Potassium Ions.
J.Mol.Biol., 436, 2024
|
|
9ETK
 
 | |
9ETG
 
 | | Crystal structure of recombinant chicken liver Bile Acid Binding Protein (cL-BABP) in complex with CA-M11 | | Descriptor: | Fatty acid-binding protein, liver, [4-[(2-azanyl-4-oxidanylidene-1,3-thiazol-5-yl)methyl]phenyl] (4~{R})-4-[(3~{R},5~{R},7~{R},8~{R},9~{S},10~{S},12~{S},13~{R},14~{S},17~{R})-10,13-dimethyl-3,7,12-tris(oxidanyl)-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1~{H}-cyclopenta[a]phenanthren-17-yl]pentanoate | | Authors: | Tassone, G, Pozzi, C, Maramai, S. | | Deposit date: | 2024-03-26 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Exploiting the bile acid binding protein as transporter of a Cholic Acid/Mirin bioconjugate for potential applications in liver cancer therapy.
Sci Rep, 14, 2024
|
|
9ETF
 
 | | Crystal structure of recombinant chicken liver Bile Acid Binding Protein (cL-BABP) in complex with lithocholic acid | | Descriptor: | (3beta,5beta,14beta,17alpha)-3-hydroxycholan-24-oic acid, Fatty acid-binding protein, liver | | Authors: | Tassone, G, Pozzi, C. | | Deposit date: | 2024-03-26 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Exploiting the bile acid binding protein as transporter of a Cholic Acid/Mirin bioconjugate for potential applications in liver cancer therapy.
Sci Rep, 14, 2024
|
|
9ETE
 
 | | Crystal structure of recombinant chicken liver Bile Acid Binding Protein (cL-BABP) in complex with deoxycholic acid | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, Fatty acid-binding protein, liver | | Authors: | Tassone, G, Pozzi, C. | | Deposit date: | 2024-03-26 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Exploiting the bile acid binding protein as transporter of a Cholic Acid/Mirin bioconjugate for potential applications in liver cancer therapy.
Sci Rep, 14, 2024
|
|
9ETD
 
 | |
9ETC
 
 | |
9EST
 
 | | STRUCTURAL STUDY OF PORCINE PANCREATIC ELASTASE COMPLEXED WITH 7-AMINO-3-(2-BROMOETHOXY)-4-CHLOROISOCOUMARIN AS A NONREACTIVATABLE DOUBLY COVALENT ENZYME-INHIBITOR COMPLEX | | Descriptor: | (2-BROMOETHYL)(2-'FORMYL-4'-AMINOPHENYL) ACETATE, CALCIUM ION, PORCINE PANCREATIC ELASTASE, ... | | Authors: | Radhakrishnan, R, Powers, J.C, Meyer Jr, E.F. | | Deposit date: | 1991-01-14 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural study of porcine pancreatic elastase complexed with 7-amino-3-(2-bromoethoxy)-4-chloroisocoumarin as a nonreactivatable doubly covalent enzyme-inhibitor complex.
Biochemistry, 30, 1991
|
|
9ESM
 
 | |
9ESA
 
 | | Aurora-C with SER mutation in complex with INCENP peptide | | Descriptor: | 1,2-ETHANEDIOL, Aurora kinase C, Inner centromere protein | | Authors: | Hillig, R.C. | | Deposit date: | 2024-03-26 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Surface-mutagenesis strategies to enable structural biology crystallization platforms.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
9ES9
 
 | | Cryo-EM structure of Spinacia oleracea cytochrome b6f complex with inhibitor DBMIB bound at plastoquinol oxidation site | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, 2,5-DIBROMO-3-ISOPROPYL-6-METHYLBENZO-1,4-QUINONE, ... | | Authors: | Pietras, R, Pintscher, S, Mielecki, B, Szwalec, M, Wojcik-Augustyn, A, Indyka, P, Rawski, M, Koziej, L, Jaciuk, M, Wazny, G, Glatt, S, Osyczka, A. | | Deposit date: | 2024-03-25 | | Release date: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.33 Å) | | Cite: | Molecular basis of plastoquinone reduction in plant cytochrome b 6 f.
Nat.Plants, 2024
|
|
9ES8
 
 | | Cryo-EM structure of Spinacia oleracea cytochrome b6f with decylplastoquinone bound at plastoquionol reduction site | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Pietras, R, Pintscher, S, Mielecki, B, Szwalec, M, Wojcik-Augustyn, A, Indyka, P, Rawski, M, Koziej, L, Jaciuk, M, Wazny, G, Glatt, S, Osyczka, A. | | Deposit date: | 2024-03-25 | | Release date: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.24 Å) | | Cite: | Molecular basis of plastoquinone reduction in plant cytochrome b 6 f.
Nat.Plants, 2024
|
|
9ES7
 
 | | Cryo-EM structure of Spinacia oleracea cytochrome b6f complex with water molecules at 1.94 A resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Pietras, R, Pintscher, S, Mielecki, B, Szwalec, M, Wojcik-Augustyn, A, Indyka, P, Rawski, M, Koziej, L, Jaciuk, M, Wazny, G, Glatt, S, Osyczka, A. | | Deposit date: | 2024-03-25 | | Release date: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (1.94 Å) | | Cite: | Molecular basis of plastoquinone reduction in plant cytochrome b 6 f.
Nat.Plants, 2024
|
|
9ERY
 
 | | Co-crystal strucutre of PD-L1 with low molecular weight inhibitor | | Descriptor: | 5-[[5-[[2-[bis(fluoranyl)methyl]-3-(2,3-dihydro-1,4-benzodioxin-6-yl)phenyl]methoxy]-2-[(2-hydroxyethylamino)methyl]phenoxy]methyl]pyridine-3-carbonitrile, Programmed cell death 1 ligand 1, SULFATE ION | | Authors: | Plewka, J, Magiera-Mularz, K, Zhang, W. | | Deposit date: | 2024-03-25 | | Release date: | 2024-07-24 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Design, synthesis, and evaluation of antitumor activity of 2-arylmethoxy-4-(2-fluoromethyl-biphenyl-3-ylmethoxy) benzylamine derivatives as PD-1/PD-l1 inhibitors.
Eur.J.Med.Chem., 276, 2024
|
|
9ERX
 
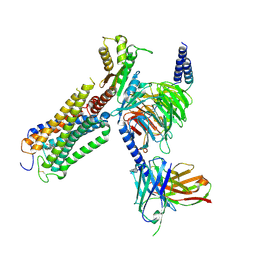 | | Structural basis of D9-THC analog activity at the Cannabinoid 1 receptor | | Descriptor: | (6aR,10aR)-9-(hydroxymethyl)-6,6-dimethyl-3-(2-methyloctan-2-yl)-6a,7,10,10a-tetrahydrobenzo[c]chromen-1-ol, Antibody ScFv16 Fab fragment, Cannabinoid receptor 1, ... | | Authors: | Thorsen, T.S, Kulkarni, Y, Boggild, A, Drace, T, Nissen, P, Gajhede, M, Boesen, T, Kastrup, J.S, Gloriam, D. | | Deposit date: | 2024-03-25 | | Release date: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of Delta 9 -THC analog activity at the Cannabinoid 1 receptor.
Res Sq, 2024
|
|
9ERO
 
 | |
9ERN
 
 | |
9ERM
 
 | |
9ER3
 
 | |
9EQP
 
 | |
9EQO
 
 | |
