8QDN
 
 | |
2ADT
 
 | | NMR structure of a 30 kDa GAAA tetraloop-receptor complex. | | Descriptor: | 43-MER | | Authors: | Davis, J.H, Tonelli, M, Scott, L.G, Jaeger, L, Williamson, J.R, Butcher, S.E. | | Deposit date: | 2005-07-20 | | Release date: | 2005-07-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | RNA Helical Packing in Solution: NMR Structure of a 30 kDa GAAA Tetraloop-Receptor Complex
J.Mol.Biol., 351, 2005
|
|
1R9V
 
 | |
1Z2F
 
 | | solution structure of CfAFP-501 | | Descriptor: | Antifreeze Protein Isoform 501 | | Authors: | Li, C, Jin, C. | | Deposit date: | 2005-03-08 | | Release date: | 2005-10-11 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of an Antifreeze Protein CfAFP-501 from Choristoneura fumiferana
J.Biomol.Nmr, 32, 2005
|
|
6NAN
 
 | | NMR structure determination of Ixolaris and Factor X interaction reveals a noncanonical mechanism of Kunitz inhibition | | Descriptor: | Ixolaris | | Authors: | De Paula, V.S, Sgourakis, N.G, Francischetti, I.M.B, Almeida, F.C.L, Monteiro, R.Q, Valente, A.P. | | Deposit date: | 2018-12-06 | | Release date: | 2019-06-12 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | NMR structure determination of Ixolaris and factor X(a) interaction reveals a noncanonical mechanism of Kunitz inhibition.
Blood, 134, 2019
|
|
1G90
 
 | |
8PIY
 
 | |
8QDP
 
 | |
8QE7
 
 | |
8QDT
 
 | |
8QEI
 
 | |
8QDK
 
 | |
8QEJ
 
 | |
8QDW
 
 | |
8QE6
 
 | |
1R2N
 
 | | NMR structure of the all-trans retinal in dark-adapted Bacteriorhodopsin | | Descriptor: | Bacteriorhodopsin, RETINAL | | Authors: | Patzelt, H, Simon, B, terLaak, A, Kessler, B, Kuhne, R, Schmieder, P, Oesterhaelt, D, Oschkinat, H. | | Deposit date: | 2003-09-29 | | Release date: | 2003-10-28 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | The structures of the active center in dark-adapted bacteriorhodopsin by solution-state NMR spectroscopy
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1EI0
 
 | |
1JU0
 
 | |
8PI0
 
 | |
1K3H
 
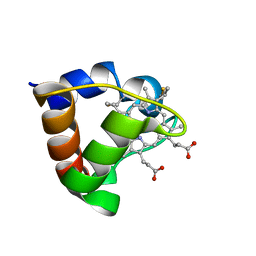 | | NMR Solution Structure of Oxidized Cytochrome c-553 from Bacillus pasteurii | | Descriptor: | HEME C, cytochrome c-553 | | Authors: | Banci, L, Bertini, I, Ciurli, S, Dikiy, A, Dittmer, J, Rosato, A, Sciara, G, Thompsett, A.R. | | Deposit date: | 2001-10-03 | | Release date: | 2001-10-31 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure, backbone mobility, and homology modeling of c-type cytochromes from gram-positive bacteria.
Chembiochem, 3, 2002
|
|
1K3G
 
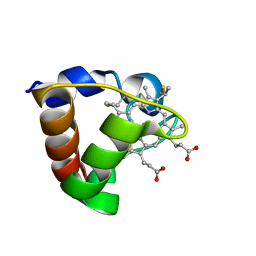 | | NMR Solution Structure of Oxidized Cytochrome c-553 from Bacillus pasteurii | | Descriptor: | HEME C, cytochrome c-553 | | Authors: | Banci, L, Bertini, I, Ciurli, S, Dikiy, A, Dittmer, J, Rosato, A, Sciara, G, Thompsett, A.R. | | Deposit date: | 2001-10-03 | | Release date: | 2001-10-31 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure, backbone mobility, and homology modeling of c-type cytochromes from gram-positive bacteria.
Chembiochem, 3, 2002
|
|
1FJK
 
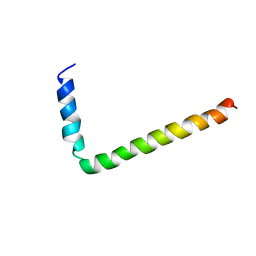 | | NMR Solution Structure of Phospholamban (C41F) | | Descriptor: | CARDIAC PHOSPHOLAMBAN | | Authors: | Lamberth, S, Griesinger, C, Schmid, H, Carafoli, E, Muenchbach, M, Vorherr, T, Krebs, J. | | Deposit date: | 2000-08-08 | | Release date: | 2000-09-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of Phospholamban
HELV.CHIM.ACTA, 83, 2000
|
|
1F8Z
 
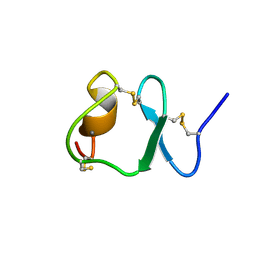 | | NMR STRUCTURE OF THE SIXTH LIGAND-BINDING MODULE OF THE LDL RECEPTOR | | Descriptor: | CALCIUM ION, LOW-DENSITY LIPOPROTEIN RECEPTOR | | Authors: | Clayton, D.J, Brereton, I.M, Kroon, P.A, Smith, R. | | Deposit date: | 2000-07-05 | | Release date: | 2000-10-18 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional NMR structure of the sixth ligand-binding module of the human LDL receptor: comparison of two adjacent modules with different ligand binding specificities.
FEBS Lett., 479, 2000
|
|
1MPZ
 
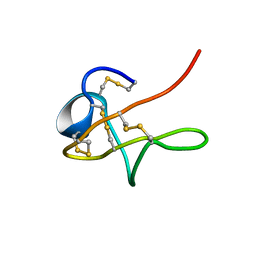 | | NMR solution structure of native Viperidae lebetina obtusa protein | | Descriptor: | Obtustatin | | Authors: | Moreno-Murciano, M.P, Monleon, D, Marcinkiewicz, C, Calvete, J.J, Celda, B. | | Deposit date: | 2002-09-13 | | Release date: | 2003-02-11 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of the Non-RGD Disintegrin Obtustatin
J.Mol.Biol., 329, 2003
|
|
1R84
 
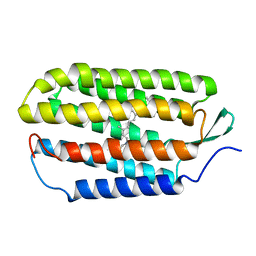 | | NMR structure of the 13-cis-15-syn retinal in dark_adapted bacteriorhodopsin | | Descriptor: | Bacteriorhodopsin, RETINAL | | Authors: | Patzelt, H, Simon, B, Ter Laak, A, Kessler, B, Kuhne, R, Schmieder, P, Oesterhaelt, D, Oschkinat, H. | | Deposit date: | 2003-10-23 | | Release date: | 2003-11-11 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | The structures of the active center in dark-adapted bacteriorhodopsin by solution-state NMR spectroscopy
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
