1B9P
 
 | |
1CP8
 
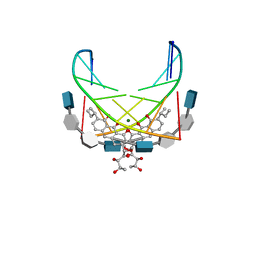 | | NMR STRUCTURE OF DNA (5'-D(TTGGCCAA)2-3') COMPLEXED WITH NOVEL ANTITUMOR DRUG UCH9 | | Descriptor: | 1,2-HYDRO-1-OXY-3,4-HYDRO-3-(1-METHOXY-2-OXY-3,4-DIHYDROXYPENTYL)-8,9-DIHYDROXY-7-(SEC-BUTYL)-ANTHRACENE, DNA (5'-D(P*TP*TP*GP*GP*CP*CP*AP*A)-3'), MAGNESIUM ION, ... | | Authors: | Katahira, R, Katahira, M, Yamashita, Y, Ogawa, H, Kyogoku, Y, Yoshida, M. | | Deposit date: | 1999-06-11 | | Release date: | 1999-07-01 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the novel antitumor drug UCH9 complexed with d(TTGGCCAA)2 as determined by NMR.
Nucleic Acids Res., 26, 1998
|
|
1AK8
 
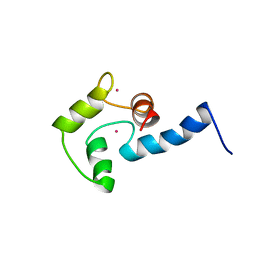 | | NMR SOLUTION STRUCTURE OF CERIUM-LOADED CALMODULIN AMINO-TERMINAL DOMAIN (CE2-TR1C), 23 STRUCTURES | | Descriptor: | CALMODULIN, CERIUM (III) ION | | Authors: | Bentrop, D, Bertini, I, Cremonini, M.A, Forsen, S, Luchinat, C, Malmendal, A. | | Deposit date: | 1997-05-29 | | Release date: | 1997-09-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the paramagnetic complex of the N-terminal domain of calmodulin with two Ce3+ ions by 1H NMR.
Biochemistry, 36, 1997
|
|
3ZGK
 
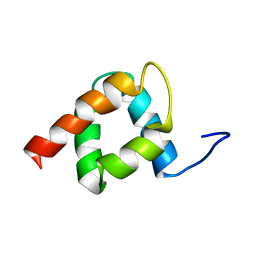 | | NMR solution structure of the RXLR effector AVR3a11 from Phytophthora Capsici | | Descriptor: | AVR3A11 | | Authors: | Tolchard, J, Chambers, V.S, Boutemy, L.S, Gathercole, R.L, Banfield, M.J, Blumenschein, T.M. | | Deposit date: | 2012-12-18 | | Release date: | 2014-01-08 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of the Avr3A11 from Phytophthora Capsi
To be Published
|
|
6ZDB
 
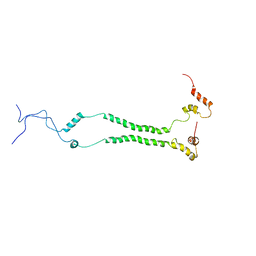 | | NMR structural analysis of yeast Cox13 reveals its C-terminus in interaction with ATP | | Descriptor: | Cytochrome c oxidase subunit 13, mitochondrial | | Authors: | Shu, Z, Pontus, P, Peter, B, Lena, M, Pia, A. | | Deposit date: | 2020-06-14 | | Release date: | 2021-05-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structural analysis of the yeast cytochrome c oxidase subunit Cox13 and its interaction with ATP.
Bmc Biol., 19, 2021
|
|
6WQR
 
 | |
5MQX
 
 | | NMR solution structure of macro domain from Venezuelan equine encephalitis virus(VEEV) in complex with ADP-ribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Non-structural protein3 | | Authors: | Makrynitsa, G.I, Ntonti, D, Marousis, K.D, Matsoukas, M.T, Papageorgiou, N, Coutard, B, Bentrop, D, Spyroulias, G.A. | | Deposit date: | 2016-12-21 | | Release date: | 2018-07-04 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Conformational plasticity of the VEEV macro domain is important for binding of ADP-ribose.
J.Struct.Biol., 206, 2019
|
|
5MN3
 
 | |
5ML1
 
 | | NMR Structure of the Littorina littorea metallothionein, a snail MT folding into three distinct domains | | Descriptor: | CADMIUM ION, Putative metallothionein | | Authors: | Baumann, C, Beil, A, Jurt, S, Zerbe, O. | | Deposit date: | 2016-12-06 | | Release date: | 2017-04-12 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural Adaptation of a Protein to Increased Metal Stress: NMR Structure of a Marine Snail Metallothionein with an Additional Domain.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5YDX
 
 | |
5YDY
 
 | |
5YIO
 
 | | NMR solution structure of subunit epsilon of the Mycobacterium tuberculosis F-ATP synthase | | Descriptor: | ATP synthase epsilon chain | | Authors: | Shin, J, Ragunathan, P, Sundararaman, L, Nartey, W, Manimekalai, M.S.S, Bogdanovic, N, Gruber, G. | | Deposit date: | 2017-10-06 | | Release date: | 2018-10-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The NMR solution structure of Mycobacterium tuberculosis F-ATP synthase subunit epsilon provides new insight into energy coupling inside the rotary engine.
FEBS J., 285, 2018
|
|
5I2V
 
 | |
8PQU
 
 | |
8QAJ
 
 | |
4CSQ
 
 | |
1GH1
 
 | | NMR STRUCTURES OF WHEAT NONSPECIFIC LIPID TRANSFER PROTEIN | | Descriptor: | NONSPECIFIC LIPID TRANSFER PROTEIN | | Authors: | Gincel, E, Simorre, J.P, Caille, A, Marion, D, Ptak, M, Vovelle, F. | | Deposit date: | 2000-10-29 | | Release date: | 2000-11-22 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure in solution of a wheat lipid-transfer protein from multidimensional 1H-NMR data. A new folding for lipid carriers.
Eur.J.Biochem., 226, 1994
|
|
7A64
 
 | |
1DK6
 
 | | NMR structure analysis of the DNA nine base pair duplex D(CATGAGTAC) D(GTAC(NP3)CATG) | | Descriptor: | 5'-D(CP*AP*TP*GP*AP*GP*TP*AP*CP*)-3', 5'-D(GP*TP*AP*CP*(NP3)P*CP*AP*TP*GP*)-3' | | Authors: | Klewer, D.A, Hoskins, A, Davisson, V.J, Bergstrom, D.E, LiWang, A.C. | | Deposit date: | 1999-12-06 | | Release date: | 2000-01-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of a DNA duplex containing nucleoside analog 1-(2'-deoxy-beta-D-ribofuranosyl)-3-nitropyrrole and the structure of the unmodified control.
Nucleic Acids Res., 28, 2000
|
|
5IX5
 
 | | NMR structure of antibacterial factor-2 | | Descriptor: | Antibacterial factor-related peptide 2 | | Authors: | Masakatsu, K, Umetsu, Y, Rumi, F, Kikukawa, T, Ohki, S, Mizuguchi, M, Demura, M, Aizawa, T. | | Deposit date: | 2016-03-23 | | Release date: | 2017-03-29 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | NMR structure of ABF-2 (antibacterial factor-2) from C. elegans and the interaction with membrane mimetic systems
To Be Published
|
|
5J6T
 
 | |
5J6V
 
 | |
5J6W
 
 | |
5U87
 
 | | NMR structure of the precursor protein PawS1 comprising SFTI-1 and a seed storage albumin | | Descriptor: | Preproalbumin PawS1 | | Authors: | Franke, B, James, A.M, Mobli, M, Colgrave, M.L, Mylne, J.S, Rosengren, K.J. | | Deposit date: | 2016-12-14 | | Release date: | 2017-03-01 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the precursor protein PawS1 comprising SFTI-1 and a seed storage albumin
to be published
|
|
5T43
 
 | |
