3G89
 
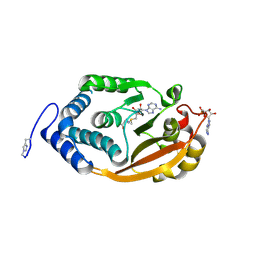 | | T. thermophilus 16S rRNA G527 methyltransferase in complex with AdoMet and AMP in space group P61 | | Descriptor: | ADENOSINE MONOPHOSPHATE, Ribosomal RNA small subunit methyltransferase G, S-ADENOSYLMETHIONINE | | Authors: | Demirci, H, Gregory, S.T, Belardinelli, R, Gualerzi, C, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2009-02-11 | | Release date: | 2009-06-30 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and functional studies of the Thermus thermophilus 16S rRNA methyltransferase RsmG
Rna, 15, 2009
|
|
3FTK
 
 | |
3FUV
 
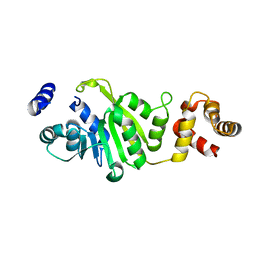 | | Apo-form of T. thermophilus 16S rRNA A1518 and A1519 methyltransferase (KsgA) in space group P43212 | | Descriptor: | Dimethyladenosine transferase | | Authors: | Demirci, H, Belardinelli, R, Seri, E, Gregory, S.T, Gualerzi, C, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2009-01-14 | | Release date: | 2009-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural rearrangements in the active site of the Thermus thermophilus 16S rRNA methyltransferase KsgA in a binary complex with 5'-methylthioadenosine.
J.Mol.Biol., 388, 2009
|
|
3G9D
 
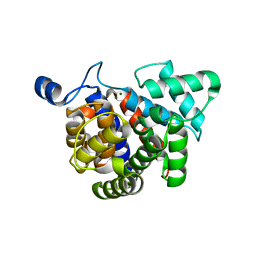 | | Crystal structure glycohydrolase | | Descriptor: | Dinitrogenase reductase activating glucohydrolase, MAGNESIUM ION | | Authors: | Li, X.-D, Winkler, F.K. | | Deposit date: | 2009-02-13 | | Release date: | 2009-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Dinitrogenase Reductase-activating Glycohydrolase (DRAG) Reveals Conservation in the ADP-Ribosylhydrolase Fold and Specific Features in the ADP-Ribose-binding Pocket
J.Mol.Biol., 390, 2009
|
|
3G9W
 
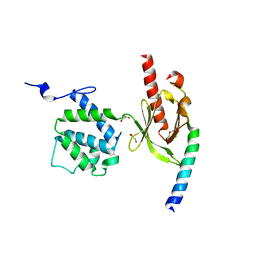 | | Crystal Structure of Talin2 F2-F3 in Complex with the Integrin Beta1D Cytoplasmic Tail | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Integrin beta-1D, ... | | Authors: | Anthis, N.J, Wegener, K.L, Ye, F, Kim, C, Lowe, E.D, Vakonakis, I, Bate, N, Critchley, D.R, Ginsberg, M.H, Campbell, I.D. | | Deposit date: | 2009-02-15 | | Release date: | 2009-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.165 Å) | | Cite: | The structure of an integrin/talin complex reveals the basis of inside-out signal transduction
Embo J., 28, 2009
|
|
3GAY
 
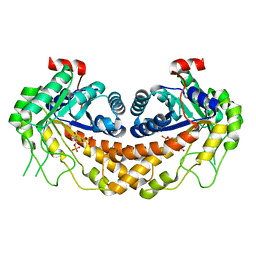 | | Structure of Giardia fructose-1,6-biphosphate aldolase in complex with tagatose-1,6-biphosphate | | Descriptor: | 1,6-di-O-phosphono-D-tagatose, Fructose-bisphosphate aldolase, ZINC ION | | Authors: | Galkin, A, Herzberg, O. | | Deposit date: | 2009-02-18 | | Release date: | 2009-03-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the substrate binding and stereoselectivity of giardia fructose-1,6-bisphosphate aldolase.
Biochemistry, 48, 2009
|
|
3G5T
 
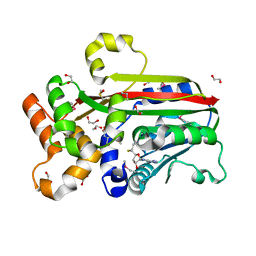 | | Crystal structure of trans-aconitate 3-methyltransferase from yeast | | Descriptor: | (2E)-2-(2-methoxy-2-oxoethyl)but-2-enedioic acid, 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, ... | | Authors: | Burgie, E.S, Bingman, C.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2009-02-05 | | Release date: | 2009-03-03 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.119 Å) | | Cite: | Crystal structure of trans-aconitate 3-methyltransferase from yeast
To be Published
|
|
3GAK
 
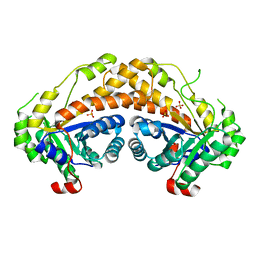 | | Structure of Giardia fructose-1,6-biphosphate aldolase | | Descriptor: | Fructose-bisphosphate aldolase, SULFATE ION, ZINC ION | | Authors: | Galkin, A, Herzberg, O. | | Deposit date: | 2009-02-17 | | Release date: | 2009-03-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural insights into the substrate binding and stereoselectivity of giardia fructose-1,6-bisphosphate aldolase.
Biochemistry, 48, 2009
|
|
3FWO
 
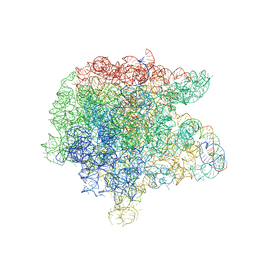 | | The large ribosomal subunit from Deinococcus radiodurans complexed with Methymycin | | Descriptor: | (3R,4S,5S,7R,9E,11S,12R)-12-ethyl-11-hydroxy-3,5,7,11-tetramethyl-2,8-dioxooxacyclododec-9-en-4-yl 3,4,6-trideoxy-3-(dimethylamino)-beta-D-xylo-hexopyranoside, 23S RIBOSOMAL RNA, 5S RIBOSOMAL RNA | | Authors: | Auerbach, T, Mermershtain, I, Bashan, A, Davidovich, C, Rozenberg, H, Sherman, D.H, Yonath, A. | | Deposit date: | 2009-01-19 | | Release date: | 2010-01-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.71 Å) | | Cite: | Structural basis for the antibacterial activity of the 12-membered-ring mono-sugar macrolide methymycin
Biotechnologia, 1, 2009
|
|
3G8A
 
 | | T. thermophilus 16S rRNA G527 methyltransferase in complex with AdoHcy in space group P61 | | Descriptor: | Ribosomal RNA small subunit methyltransferase G, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Demirci, H, Gregory, S.T, Belardinelli, R, Gualerzi, C, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2009-02-11 | | Release date: | 2009-06-30 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and functional studies of the Thermus thermophilus 16S rRNA methyltransferase RsmG
Rna, 15, 2009
|
|
3G2X
 
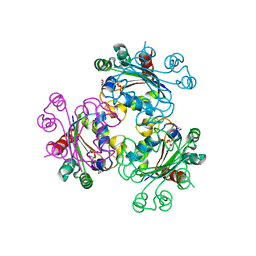 | | Structure of mimivirus NDK +Kpn - N62L double mutant complexed with dTDP | | Descriptor: | MAGNESIUM ION, Nucleoside diphosphate kinase, THYMIDINE-5'-DIPHOSPHATE | | Authors: | Jeudy, S, Lartigue, A, Claverie, J.M, Abergel, C. | | Deposit date: | 2009-02-01 | | Release date: | 2009-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Dissecting the unique nucleotide specificity of mimivirus nucleoside diphosphate kinase.
J.Virol., 83, 2009
|
|
3DKD
 
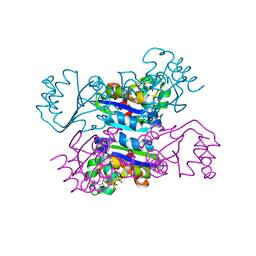 | | Crystal structure of the mimivirus NDK +Kpn-N62L-R107G triple mutant complexed with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Nucleoside diphosphate kinase | | Authors: | Jeudy, S, Lartigue, A, Claverie, J.M, Abergel, C. | | Deposit date: | 2008-06-24 | | Release date: | 2009-08-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Dissecting the unique nucleotide specificity of mimivirus nucleoside diphosphate kinase.
J.Virol., 83, 2009
|
|
3DMF
 
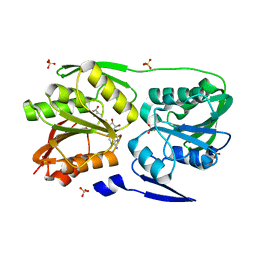 | | T. Thermophilus 16S rRNA N2 G1207 methyltransferase (RsmC) in complex with AdoMet | | Descriptor: | Probable ribosomal RNA small subunit methyltransferase, S-ADENOSYLMETHIONINE, SULFATE ION | | Authors: | Demirci, H, Gregory, S.T, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2008-07-01 | | Release date: | 2008-07-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal Structure of the Thermus thermophilus 16 S rRNA Methyltransferase RsmC in Complex with Cofactor and Substrate Guanosine.
J.Biol.Chem., 283, 2008
|
|
3G8B
 
 | | T. thermophilus 16S rRNA G527 methyltransferase in complex with AdoMet in space group I222 | | Descriptor: | Ribosomal RNA small subunit methyltransferase G, S-ADENOSYLMETHIONINE, SULFATE ION | | Authors: | Demirci, H, Gregory, S.T, Belardinelli, R, Gualerzi, C, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2009-02-11 | | Release date: | 2009-06-30 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and functional studies of the Thermus thermophilus 16S rRNA methyltransferase RsmG
Rna, 15, 2009
|
|
3G1E
 
 | | X-ray crystal structure of coil 1A of human vimentin | | Descriptor: | Vimentin | | Authors: | Meier, M, Padilla, G.P, Herrmann, H, Wedig, T, Hergt, M, Patel, T.R, Stetefeld, J, Aebi, U, Burkhard, P. | | Deposit date: | 2009-01-29 | | Release date: | 2009-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Vimentin coil 1A-A molecular switch involved in the initiation of filament elongation.
J.Mol.Biol., 390, 2009
|
|
3GB6
 
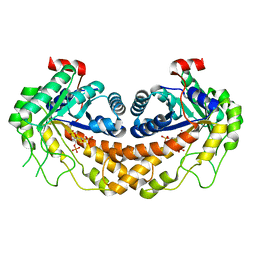 | | Structure of Giardia fructose-1,6-biphosphate aldolase D83A mutant in complex with fructose-1,6-bisphosphate | | Descriptor: | 1,6-di-O-phosphono-D-fructose, Fructose-bisphosphate aldolase, ZINC ION | | Authors: | Galkin, A, Herzberg, O. | | Deposit date: | 2009-02-18 | | Release date: | 2009-03-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into the substrate binding and stereoselectivity of giardia fructose-1,6-bisphosphate aldolase.
Biochemistry, 48, 2009
|
|
3G5P
 
 | | Structure and activity of human mitochondrial peptide deformylase, a novel cancer target | | Descriptor: | COBALT (II) ION, PHOSPHATE ION, Peptide deformylase, ... | | Authors: | Escobar-Alvarez, S, Goldgur, Y, Yang, G, Ouerfelli, O, Li, Y, Scheinberg, D.A. | | Deposit date: | 2009-02-05 | | Release date: | 2009-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and activity of human mitochondrial peptide deformylase, a novel cancer target
J.Mol.Biol., 387, 2009
|
|
2UUL
 
 | | Crystal structure of C-phycocyanin from Phormidium, Lyngbya spp. (Marine) and Spirulina sp. (Fresh water) shows two different ways of energy transfer between two hexamers. | | Descriptor: | BILIVERDINE IX ALPHA, C-PHYCOCYANIN ALPHA CHAIN, C-PHYCOCYANIN BETA CHAIN, ... | | Authors: | Satyanarayana, L, Patel, A, Mishra, S, K Ghosh, P, Suresh, C.G. | | Deposit date: | 2007-03-04 | | Release date: | 2008-05-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structure of C-Phycocyanin from Phormidium, Lyngbya Spp. (Marine) and Spirulina Sp. (Fresh Water) Shows Two Different Ways of Energy Transfer between Twohexamers.
To be Published
|
|
5Q0U
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3, trans-4-({(2S)-2-[2-(4-chlorophenyl)-5,6-difluoro-1H-benzimidazol-1-yl]-2-cyclohexylacetyl}amino)cyclohexyl hydrogen sulfate | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
3G88
 
 | | T. thermophilus 16S rRNA G527 methyltransferase in complex with AdoMet in space group P61 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Ribosomal RNA small subunit methyltransferase G, S-ADENOSYLMETHIONINE | | Authors: | Demirci, H, Belardinelli, R, Gregory, S.T, Gualerzi, C, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2009-02-11 | | Release date: | 2009-06-30 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural and functional studies of the Thermus thermophilus 16S rRNA methyltransferase RsmG
Rna, 15, 2009
|
|
3GAN
 
 | | Crystal structure of gene product from Arabidopsis thaliana At3g22680 with bound suramin | | Descriptor: | 8,8'-[CARBONYLBIS[IMINO-3,1-PHENYLENECARBONYLIMINO(4-METHYL-3,1-PHENYLENE)CARBONYLIMINO]]BIS-1,3,5-NAPHTHALENETRISULFON IC ACID, CHLORIDE ION, Uncharacterized protein At3g22680 | | Authors: | Burgie, E.S, Bingman, C.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2009-02-17 | | Release date: | 2009-03-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of gene product from Arabidopsis thaliana At3g22680 with bound suramin
To be Published
|
|
1HEU
 
 | | ATOMIC X-RAY STRUCTURE OF LIVER ALCOHOL DEHYDROGENASE CONTAINING Cadmium and a hydroxide adduct to NADH | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, ALCOHOL DEHYDROGENASE E CHAIN, CADMIUM ION, ... | | Authors: | Meijers, R, Morris, R.J, Adolph, H.W, Merli, A, Lamzin, V.S, Cedergen-Zeppezauer, E.S. | | Deposit date: | 2000-11-26 | | Release date: | 2001-05-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | On the Enzymatic Activation of Nadh
J.Biol.Chem., 276, 2001
|
|
3EJ6
 
 | | Neurospora Crassa Catalase-3 Crystal Structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Catalase-3, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Diaz, A, Valdes, V.-J, Rudino-Pinera, E, Horjales, E, Hansberg, W. | | Deposit date: | 2008-09-17 | | Release date: | 2009-01-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Function Relationships in Fungal Large-Subunit Catalases
J.Mol.Biol., 386, 2008
|
|
1HF3
 
 | | ATOMIC X-RAY STRUCTURE OF LIVER ALCOHOL DEHYDROGENASE CONTAINING Cadmium and a hydroxide adduct to NADH | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, ALCOHOL DEHYDROGENASE E CHAIN, CADMIUM ION, ... | | Authors: | Meijers, R, Morris, R.J, Adolph, H.W, Merli, A, Lamzin, V.S, Cedergen-Zeppezauer, E.S. | | Deposit date: | 2000-11-27 | | Release date: | 2001-05-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | On the Enzymatic Activation of Nadh
J.Biol.Chem., 276, 2001
|
|
3EBD
 
 | | Structure of inhibited murine iNOS oxygenase domain | | Descriptor: | (2S)-2-methyl-2,3-dihydrothieno[2,3-f][1,4]oxazepin-5-amine, 5,6,7,8-TETRAHYDROBIOPTERIN, Nitric oxide synthase, ... | | Authors: | Garcin, E.D, Arvai, A.S, Rosenfeld, R.J, Kroeger, M.D, Crane, B.R, Andersson, G, Andrews, G, Hamley, P.J, Mallinder, P.R, Nicholls, D.J, St-Gallay, S.A, Tinker, A.C, Gensmantel, N.P, Mete, A, Cheshire, D.R, Connolly, S, Stuehr, D.J, Aberg, A, Wallace, A.V, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2008-08-27 | | Release date: | 2008-10-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Anchored plasticity opens doors for selective inhibitor design in nitric oxide synthase.
Nat.Chem.Biol., 4, 2008
|
|
