6VAJ
 
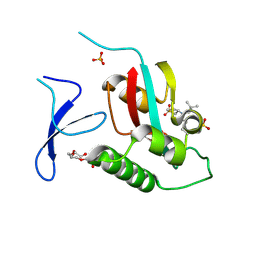 | | Crystal Structure Analysis of human PIN1 | | Descriptor: | 2-chloro-N-(2,2-dimethylpropyl)-N-[(3R)-1,1-dioxo-1lambda~6~-thiolan-3-yl]acetamide, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, SULFATE ION, ... | | Authors: | Seo, H.-S, Dhe-Paganon, S. | | Deposit date: | 2019-12-17 | | Release date: | 2020-12-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Sulfopin is a covalent inhibitor of Pin1 that blocks Myc-driven tumors in vivo.
Nat.Chem.Biol., 17, 2021
|
|
2QJC
 
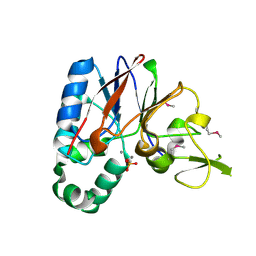 | | Crystal structure of a putative diadenosine tetraphosphatase | | Descriptor: | Diadenosine tetraphosphatase, putative, MANGANESE (II) ION, ... | | Authors: | Sugadev, R, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-07-06 | | Release date: | 2007-07-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural genomics of protein phosphatases.
J.Struct.Funct.Genom., 8, 2007
|
|
6MDC
 
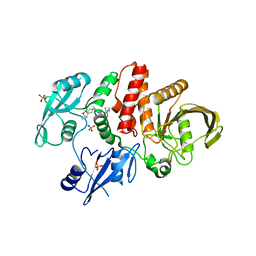 | | Non-receptor Protein Tyrosine Phosphatase SHP2 in Complex with Allosteric Inhibitor Pyrazolo-pyrimidinone 1 SHP389 | | Descriptor: | 6-[(3S,4S)-4-amino-3-methyl-2-oxa-8-azaspiro[4.5]decan-8-yl]-3-[3-chloro-2-(cyclopropylamino)pyridin-4-yl]-5-methyl-2,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, PHOSPHATE ION, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Fodor, M, Stams, T. | | Deposit date: | 2018-09-04 | | Release date: | 2019-02-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Optimization of Fused Bicyclic Allosteric SHP2 Inhibitors.
J. Med. Chem., 62, 2019
|
|
7HCV
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with AVI-0000610 | | Descriptor: | (3R,5R)-5-(hydroxymethyl)-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)pyrrolidin-3-ol, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2024-08-15 | | Release date: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Exploration of structure-activity relationships for the SARS-CoV-2 macrodomain from shape-based fragment linking and active learning.
Sci Adv, 11, 2025
|
|
4OOV
 
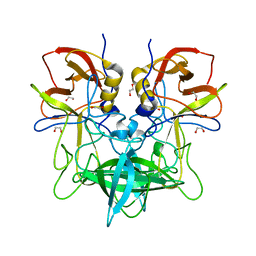 | |
7HCO
 
 | |
7HD8
 
 | |
7HCZ
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with AVI-0003702 | | Descriptor: | CHLORIDE ION, Non-structural protein 3, [(8R)-7-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)-1,4-dioxa-7-azaspiro[4.4]nonan-8-yl]methanol, ... | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2024-08-15 | | Release date: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Exploration of structure-activity relationships for the SARS-CoV-2 macrodomain from shape-based fragment linking and active learning.
Sci Adv, 11, 2025
|
|
7HDO
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with AVI-0003637 | | Descriptor: | (2S)-4,4,4-trifluoro-2-[(7H-pyrrolo[2,3-d]pyrimidin-4-yl)amino]butan-1-ol, CHLORIDE ION, Non-structural protein 3, ... | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2024-08-15 | | Release date: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Exploration of structure-activity relationships for the SARS-CoV-2 macrodomain from shape-based fragment linking and active learning.
Sci Adv, 11, 2025
|
|
7HEP
 
 | |
5VVE
 
 | |
3H36
 
 | | Structure of an uncharacterized domain in polyribonucleotide nucleotidyltransferase from Streptococcus mutans UA159 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Polyribonucleotide nucleotidyltransferase | | Authors: | Cuff, M.E, Hatzos, C, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-04-15 | | Release date: | 2009-05-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of an uncharacterized domain in polyribonucleotide nucleotidyltransferase from Streptococcus mutans UA159
TO BE PUBLISHED
|
|
7HDX
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with AVI-0003648 | | Descriptor: | (2S)-4,4-dimethyl-2-[(7H-pyrrolo[2,3-d]pyrimidin-4-yl)amino]pentan-1-ol, CHLORIDE ION, Non-structural protein 3, ... | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2024-08-15 | | Release date: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Exploration of structure-activity relationships for the SARS-CoV-2 macrodomain from shape-based fragment linking and active learning.
Sci Adv, 11, 2025
|
|
3DTW
 
 | | Crystal structure of the VEGFR2 kinase domain in complex with a benzisoxazole inhibitor | | Descriptor: | 6-chloro-N-pyrimidin-5-yl-3-{[3-(trifluoromethyl)phenyl]amino}-1,2-benzisoxazole-7-carboxamide, Vascular endothelial growth factor receptor 2 | | Authors: | Whittington, D.A, Long, A.M, Rose, P, Gu, Y, Zhao, H. | | Deposit date: | 2008-07-16 | | Release date: | 2008-09-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of amido-benzisoxazoles as potent c-Kit inhibitors.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
7HD2
 
 | |
7HDN
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with AVI-0003734 | | Descriptor: | (2R)-2-[(7H-pyrrolo[2,3-d]pyrimidin-4-yl)amino]butane-1,4-diol, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2024-08-15 | | Release date: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Exploration of structure-activity relationships for the SARS-CoV-2 macrodomain from shape-based fragment linking and active learning.
Sci Adv, 11, 2025
|
|
5VH6
 
 | | 2.6 Angstrom Resolution Crystal Structure of N-terminal Fragment (residues 1-406) of Elongation Factor G from Bacillus subtilis. | | Descriptor: | CHLORIDE ION, Elongation factor G | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-04-12 | | Release date: | 2017-04-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | 2.6 Angstrom Resolution Crystal Structure of N-terminal Fragment (residues 1-406) of Elongation Factor G from Bacillus subtilis.
To Be Published
|
|
6UYH
 
 | |
7HDU
 
 | |
7HF5
 
 | |
7HF7
 
 | |
5COE
 
 | | The structure of the NK1 fragment of HGF/SF complexed with HEPES | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Hepatocyte growth factor | | Authors: | Sigurdardottir, A.G, Winter, A, Sobkowicz, A, Fragai, M, Ascher, D.B, Chirgadze, D.Y, Blundell, T.L, Gherardi, E. | | Deposit date: | 2015-07-20 | | Release date: | 2015-08-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Exploring the chemical space of the lysine-binding pocket of the first kringle domain of hepatocyte growth factor/scatter factor (HGF/SF) yields a new class of inhibitors of HGF/SF-MET binding.
Chem Sci, 6, 2015
|
|
7HED
 
 | |
7HF8
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with AVI-0004329 | | Descriptor: | (1r,4r)-1-{[(5-methyl-7H-pyrrolo[2,3-d]pyrimidin-4-yl)amino]methyl}cyclohexane-1,4-diol, CHLORIDE ION, Non-structural protein 3 | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2024-08-15 | | Release date: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | Exploration of structure-activity relationships for the SARS-CoV-2 macrodomain from shape-based fragment linking and active learning.
Sci Adv, 11, 2025
|
|
4O9E
 
 | | Crystal structure of QdtA, a sugar 3,4-ketoisemerase from Thermoanaerobacterium thermosaccharolyticum in complex with TDP | | Descriptor: | (2S)-1-[3-[(2S)-2-oxidanylpropoxy]-2-[[(2S)-2-oxidanylpropoxy]methyl]-2-[[(2R)-2-oxidanylpropoxy]methyl]propoxy]propan-2-ol, QdtA, THYMIDINE-5'-DIPHOSPHATE, ... | | Authors: | Thoden, J.B, Holden, H.M. | | Deposit date: | 2014-01-02 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The molecular architecture of QdtA, a sugar 3,4-ketoisomerase from Thermoanaerobacterium thermosaccharolyticum.
Protein Sci., 23, 2014
|
|
