2XUC
 
 | | Natural product-guided discovery of a fungal chitinase inhibitor | | Descriptor: | 1-methyl-3-(N-methylcarbamimidoyl)urea, CHITINASE, CHLORIDE ION, ... | | Authors: | Rush, C.L, Schuttelkopf, A.W, Hurtado-Guerrero, R, Blair, D.E, Ibrahim, A.F.M, Desvergnes, S, Eggleston, I.M, van Aalten, D.M.F. | | Deposit date: | 2010-10-18 | | Release date: | 2010-10-27 | | Last modified: | 2019-01-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Natural Product-Guided Discovery of a Fungal Chitinase Inhibitor.
Chem.Biol., 17, 2010
|
|
3EBK
 
 | | Crystal structure of major allergens, Bla g 4 from cockroaches | | Descriptor: | Allergen Bla g 4 | | Authors: | Tan, Y.W, Chan, S.L, Chew, F.T, Sivaraman, J, Mok, Y.K. | | Deposit date: | 2008-08-28 | | Release date: | 2008-12-02 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of two major allergens, Bla g 4 and Per a 4, from cockroaches and their IgE binding epitopes.
J.Biol.Chem., 284, 2008
|
|
8IYB
 
 | | Structure insight into substrate recognition and catalysis by feruloyl esterase from Aspergillus sydowii | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, ... | | Authors: | Phienluphon, A, Kondo, K, Mikami, B, Nagata, T, Katahira, M. | | Deposit date: | 2023-04-04 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insights into the molecular mechanisms of substrate recognition and hydrolysis by feruloyl esterase from Aspergillus sydowii.
Int.J.Biol.Macromol., 253, 2023
|
|
8IYC
 
 | | Structure insight into substrate recognition and catalysis by feruloyl esterase from Aspergillus sydowii | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Phienluphon, A, Kondo, K, Mikami, B, Nagata, T, Katahira, M. | | Deposit date: | 2023-04-04 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural insights into the molecular mechanisms of substrate recognition and hydrolysis by feruloyl esterase from Aspergillus sydowii.
Int.J.Biol.Macromol., 253, 2023
|
|
8IY8
 
 | | Structure insight into substrate recognition and catalysis by feruloyl esterase from Aspergillus sydowii | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Feruloyl esterase | | Authors: | Phienluphon, A, Kondo, K, Mikami, B, Nagata, T, Katahira, M. | | Deposit date: | 2023-04-04 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insights into the molecular mechanisms of substrate recognition and hydrolysis by feruloyl esterase from Aspergillus sydowii.
Int.J.Biol.Macromol., 253, 2023
|
|
1LBS
 
 | | LIPASE (E.C.3.1.1.3) (TRIACYLGLYCEROL HYDROLASE) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LIPASE B, N-HEXYLPHOSPHONATE ETHYL ESTER | | Authors: | Uppenberg, J, Jones, T.A. | | Deposit date: | 1995-07-11 | | Release date: | 1995-10-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystallographic and molecular-modeling studies of lipase B from Candida antarctica reveal a stereospecificity pocket for secondary alcohols.
Biochemistry, 34, 1995
|
|
3GLY
 
 | | REFINED CRYSTAL STRUCTURES OF GLUCOAMYLASE FROM ASPERGILLUS AWAMORI VAR. X100 | | Descriptor: | GLUCOAMYLASE-471, alpha-D-mannopyranose, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Aleshin, A.E, Hoffman, C, Firsov, L.M, Honzatko, R.B. | | Deposit date: | 1994-06-03 | | Release date: | 1994-11-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Refined crystal structures of glucoamylase from Aspergillus awamori var. X100.
J.Mol.Biol., 238, 1994
|
|
2E4O
 
 | | X-ray Crystal Structure of Aristolochene Synthase from Aspergillus terreus and the Evolution of Templates for the Cyclization of Farnesyl Diphosphate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Aristolochene synthase, BETA-MERCAPTOETHANOL, ... | | Authors: | Shishova, E.Y, Di Costanzo, L, Cane, D.E, Christianson, D.W. | | Deposit date: | 2006-12-15 | | Release date: | 2007-01-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray crystal structure of aristolochene synthase from Aspergillus terreus and evolution of templates for the cyclization of farnesyl diphosphate.
Biochemistry, 46, 2007
|
|
1LBT
 
 | | LIPASE (E.C.3.1.1.3) (TRIACYLGLYCEROL HYDROLASE) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LIPASE B, METHYLPENTA(OXYETHYL) HEPTADECANOATE | | Authors: | Uppenberg, J, Jones, T.A. | | Deposit date: | 1995-07-11 | | Release date: | 1995-10-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallographic and molecular-modeling studies of lipase B from Candida antarctica reveal a stereospecificity pocket for secondary alcohols.
Biochemistry, 34, 1995
|
|
1Y4W
 
 | | Crystal structure of exo-inulinase from Aspergillus awamori in spacegroup P21 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Nagem, R.A.P, Rojas, A.L, Golubev, A.M, Korneeva, O.S, Eneyskaya, E.V, Kulminskaya, A.A, Neustroev, K.N, Polikarpov, I. | | Deposit date: | 2004-12-01 | | Release date: | 2004-12-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of exo-inulinase from Aspergillus awamori: the enzyme fold and structural determinants of substrate recognition
J.Mol.Biol., 344, 2004
|
|
1KTJ
 
 | | X-ray Structure Of Der P 2, The Major House Dust Mite Allergen | | Descriptor: | ALLERGEN DER P 2 | | Authors: | Derewenda, U, Li, J, Derewenda, Z, Dauter, Z, Mueller, G.A, Rule, G.S, Benjamin, D.C. | | Deposit date: | 2002-01-16 | | Release date: | 2002-05-15 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The crystal structure of a major dust mite allergen Der p 2, and its biological implications.
J.Mol.Biol., 318, 2002
|
|
2X8R
 
 | | The structure of a family GH25 lysozyme from Aspergillus fumigatus | | Descriptor: | CHLORIDE ION, GLYCOSYL HYDROLASE | | Authors: | Korczynska, J.E, Danielsen, S, Schagerlof, U, Turkenburg, J.P, Davies, G.J, Wilson, K.S, Taylor, E.J. | | Deposit date: | 2010-03-11 | | Release date: | 2010-09-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Structure of a Family Gh25 Lysozyme from Aspergillus Fumigatus
Acta Crystallogr.,Sect.F, 66, 2010
|
|
5VYL
 
 | |
5I78
 
 | | Crystal structure of a beta-1,4-endoglucanase from Aspergillus niger | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Endo-beta-1, ... | | Authors: | Liu, W.D, Yan, J.J, Li, Y.J, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2016-02-17 | | Release date: | 2016-12-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Functional and structural analysis of Pichia pastoris-expressed Aspergillus niger 1,4-beta-endoglucanase
Biochem. Biophys. Res. Commun., 475, 2016
|
|
5I79
 
 | | Crystal structure of a beta-1,4-endoglucanase mutant from Aspergillus niger in complex with sugar | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Endo-beta-1, ... | | Authors: | Liu, W.D, Yan, J.J, Li, Y.J, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2016-02-17 | | Release date: | 2016-12-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Functional and structural analysis of Pichia pastoris-expressed Aspergillus niger 1,4-beta-endoglucanase
Biochem. Biophys. Res. Commun., 475, 2016
|
|
5I77
 
 | | Crystal structure of a beta-1,4-endoglucanase from Aspergillus niger | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Li, Y.J, Liu, W.D, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2016-02-17 | | Release date: | 2016-12-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Functional and structural analysis of Pichia pastoris-expressed Aspergillus niger 1,4-beta-endoglucanase
Biochem. Biophys. Res. Commun., 475, 2016
|
|
3G02
 
 | |
3BLJ
 
 | | Crystal structure of human poly(ADP-ribose) polymerase 15, catalytic fragment | | Descriptor: | CHLORIDE ION, GLYCEROL, Poly(ADP-ribose) polymerase 15, ... | | Authors: | Karlberg, T, Lehtio, L, Arrowsmith, C.H, Berglund, H, Busam, R.D, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Kallas, A, Kotenyova, T, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Svensson, L, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Welin, M, Weigelt, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-12-11 | | Release date: | 2007-12-25 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for Lack of ADP-ribosyltransferase Activity in Poly(ADP-ribose) Polymerase-13/Zinc Finger Antiviral Protein.
J.Biol.Chem., 290, 2015
|
|
5MYI
 
 | | Convergent evolution involving dimeric and trimeric dUTPases in signalling. | | Descriptor: | CALCIUM ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Donderis, J, Bowring, J, Maiques, E, Ciges-Tomas, J.R, Alite, C, Mehmedov, I, Tormo-Mas, M.A, Penades, J.R, Marina, A. | | Deposit date: | 2017-01-26 | | Release date: | 2017-09-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Convergent evolution involving dimeric and trimeric dUTPases in pathogenicity island mobilization.
PLoS Pathog., 13, 2017
|
|
5MYD
 
 | | Convergent evolution involving dimeric and trimeric dUTPases in signalling. | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, DUTPase, MAGNESIUM ION | | Authors: | Donderis, J, Bowring, J, Maiques, E, Ciges-Tomas, J.R, Alite, C, Mehmedov, I, Tormo-Mas, M.A, Penades, J.R, Marina, A. | | Deposit date: | 2017-01-26 | | Release date: | 2017-09-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Convergent evolution involving dimeric and trimeric dUTPases in pathogenicity island mobilization.
PLoS Pathog., 13, 2017
|
|
2C3B
 
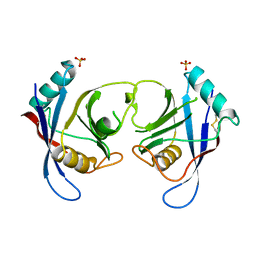 | | The Crystal Structure of Aspergillus fumigatus Cyclophilin reveals 3D Domain Swapping of a Central Element | | Descriptor: | PPIASE, SULFATE ION | | Authors: | Limacher, A, Kloer, D.P, Fluckiger, S, Folkers, G, Crameri, R, Scapozza, L. | | Deposit date: | 2005-10-05 | | Release date: | 2006-01-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Crystal Structure of Aspergillus Fumigatus Cyclophilin Reveals 3D Domain Swapping of a Central Element
Structure, 14, 2006
|
|
3G0I
 
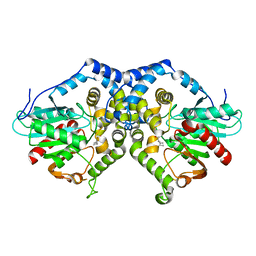 | |
5MYF
 
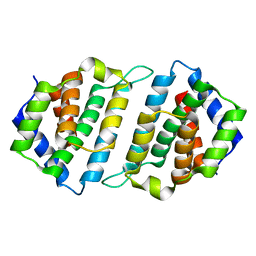 | | Convergent evolution involving dimeric and trimeric dUTPases in signalling. | | Descriptor: | dUTPase from DI S. aureus phage | | Authors: | Donderis, J, Bowring, J, Maiques, E, Ciges-Tomas, J.R, Alite, C, Mehmedov, I, Tormo-Mas, M.A, Penades, J.R, Marina, A. | | Deposit date: | 2017-01-26 | | Release date: | 2017-09-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Convergent evolution involving dimeric and trimeric dUTPases in pathogenicity island mobilization.
PLoS Pathog., 13, 2017
|
|
1N0V
 
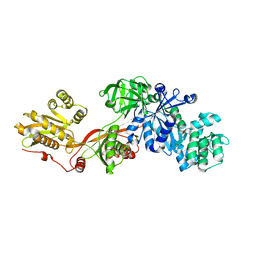 | | Crystal structure of elongation factor 2 | | Descriptor: | Elongation factor 2 | | Authors: | Joergensen, R, Ortiz, P.A, Carr-Schmid, A, Nissen, P, Kinzy, T.G, Andersen, G.R. | | Deposit date: | 2002-10-15 | | Release date: | 2002-11-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Two crystal structures demonstrate large conformational changes in the eukaryotic ribosomal translocase.
Nat.Struct.Biol., 10, 2003
|
|
5YOT
 
 | | Isoprimeverose-producing enzyme from Aspergillus oryzae in complex with isoprimeverose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Matsuzawa, T, Watanabe, M, Nakamichi, Y, Yaoi, K. | | Deposit date: | 2017-10-31 | | Release date: | 2018-11-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal structure and substrate recognition mechanism of Aspergillus oryzae isoprimeverose-producing enzyme.
J.Struct.Biol., 205, 2019
|
|
