1MFG
 
 | |
1MFL
 
 | |
6J8Y
 
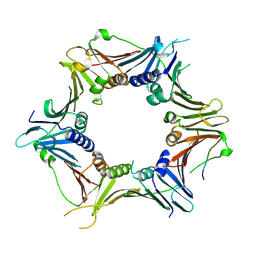 | | Crystal structure of the human RAD9-HUS1-RAD1-RHINO complex | | Descriptor: | Cell cycle checkpoint control protein RAD9A, Cell cycle checkpoint protein RAD1, Checkpoint protein HUS1, ... | | Authors: | Hara, K, Iida, N, Sakurai, H, Hashimoto, H. | | Deposit date: | 2019-01-21 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the RAD9-RAD1-HUS1 checkpoint clamp bound to RHINO sheds light on the other side of the DNA clamp.
J.Biol.Chem., 295, 2020
|
|
7EAL
 
 | |
6XAV
 
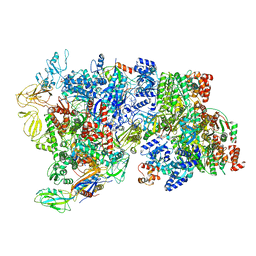 | | CryoEM Structure of E. coli Rho-dependent Transcription Pre-termination Complex bound with NusG | | Descriptor: | DNA (29-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Hao, Z.T, Kim, H.K, Walz, T, Nudler, E. | | Deposit date: | 2020-06-04 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Pre-termination Transcription Complex: Structure and Function.
Mol.Cell, 81, 2021
|
|
7EAO
 
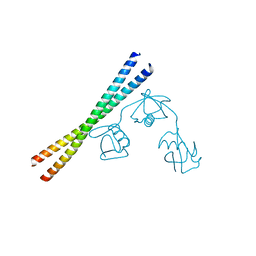 | |
7FCZ
 
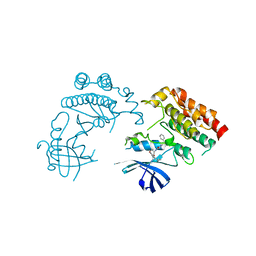 | | Crystal Structure of human RIPK1 kinase domain in complex with a novel inhibitor | | Descriptor: | N-[(3S)-7-(2-cyclopropylethynyl)-5-methyl-4-oxidanylidene-2,3-dihydro-1,5-benzoxazepin-3-yl]-5-(phenylmethyl)-4H-1,2,4-triazole-3-carboxamide, Receptor-interacting serine/threonine-protein kinase 1 | | Authors: | Su, H.X, Xie, H, Nie, T.Q, Li, M.J, Xu, Y.C. | | Deposit date: | 2021-07-15 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Potent and Selective RIPK1 Inhibitors Targeting Dual-Pockets for the Treatment of Systemic Inflammatory Response Syndrome and Sepsis.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7FD0
 
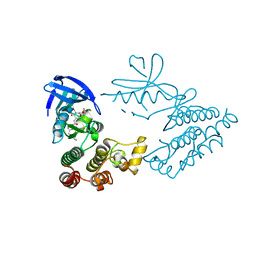 | | Crystal Structure of human RIPK1 kinase domain in complex with a novel inhibitor | | Descriptor: | N-[(3S)-5-methyl-7-[2-(oxan-4-yl)ethynyl]-4-oxidanylidene-2,3-dihydro-1,5-benzoxazepin-3-yl]-5-(phenylmethyl)-4H-1,2,4-triazole-3-carboxamide, Receptor-interacting serine/threonine-protein kinase 1 | | Authors: | Su, H.X, Xie, H, Nie, T.Q, Li, M.J, Xu, Y.C. | | Deposit date: | 2021-07-15 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Potent and Selective RIPK1 Inhibitors Targeting Dual-Pockets for the Treatment of Systemic Inflammatory Response Syndrome and Sepsis.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
5L1P
 
 | | X-ray Structure of Cytochrome P450 PntM with Pentalenolactone | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Pentalenolactone synthase, pentalenolactone | | Authors: | Duan, L, Jogl, G, Cane, D.E. | | Deposit date: | 2016-07-29 | | Release date: | 2016-09-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | The Cytochrome P450-Catalyzed Oxidative Rearrangement in the Final Step of Pentalenolactone Biosynthesis: Substrate Structure Determines Mechanism.
J.Am.Chem.Soc., 138, 2016
|
|
4KC7
 
 | | Crystal Structure of Endo-1,5-alpha-L-arabinanase from Thermotoga petrophila RKU-1 | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, Glycoside hydrolase, ... | | Authors: | Nascimento, A.F.Z, Polo, C.C, Santos, C.R, Costa, M.C.M.F, Mesa, A.N, Prade, R.A, Ruller, R, Squina, F.M, Murakami, M.T. | | Deposit date: | 2013-04-24 | | Release date: | 2014-02-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Mechanistic strategies for catalysis adopted by evolutionary distinct family 43 arabinanases.
J.Biol.Chem., 289, 2014
|
|
4KCB
 
 | | Crystal Structure of Exo-1,5-alpha-L-arabinanase from Bovine Ruminal Metagenomic Library | | Descriptor: | Arabinan endo-1,5-alpha-L-arabinosidase, PHOSPHATE ION | | Authors: | Santos, C.R, Polo, C.C, Costa, M.C.M.F, Nascimento, A.F.Z, Wong, D.W.S, Murakami, M.T. | | Deposit date: | 2013-04-24 | | Release date: | 2014-02-05 | | Last modified: | 2014-04-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mechanistic strategies for catalysis adopted by evolutionary distinct family 43 arabinanases.
J.Biol.Chem., 289, 2014
|
|
6ZE5
 
 | | FAD-dependent oxidoreductase from Chaetomium thermophilum in complex with fragment 2-(1H-indol-3-yl)-N-[(1-methyl-1H-pyrrol-2-yl)methyl]ethanamine | | Descriptor: | 2-(1H-indol-3-yl)-N-[(1-methyl-1H-pyrrol-2-yl)methyl]ethanamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Svecova, L, Skalova, T, Kolenko, P, Koval, T, Oestergaard, L.H, Dohnalek, J. | | Deposit date: | 2020-06-16 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystallographic fragment screening-based study of a novel FAD-dependent oxidoreductase from Chaetomium thermophilum.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
6ZE6
 
 | | FAD-dependent oxidoreductase from Chaetomium thermophilum in complex with fragment 4-nitrocatechol | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-NITROCATECHOL, ... | | Authors: | Svecova, L, Skalova, T, Kolenko, P, Koval, T, Oestergaard, L.H, Dohnalek, J. | | Deposit date: | 2020-06-16 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Crystallographic fragment screening-based study of a novel FAD-dependent oxidoreductase from Chaetomium thermophilum.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
6ZE7
 
 | | Chaetomium thermophilum FAD-dependent oxidoreductase in complex with 4-nitrophenol | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Svecova, L, Skalova, T, Kolenko, P, Koval, T, Oestergaard, L.H, Dohnalek, J. | | Deposit date: | 2020-06-16 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystallographic fragment screening-based study of a novel FAD-dependent oxidoreductase from Chaetomium thermophilum.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
6ZE4
 
 | | FAD-dependent oxidoreductase from Chaetomium thermophilum in complex with fragment 4-oxo-N-[(1S)-1-(pyridin-3-yl)ethyl]-4-(thiophen-2-yl)butanamide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-oxo-N-[(1S)-1-(pyridin-3-yl)ethyl]-4-(thiophen-2-yl)butanamide, ... | | Authors: | Svecova, L, Skalova, T, Kolenko, P, Koval, T, Oestergaard, L.H, Dohnalek, J. | | Deposit date: | 2020-06-16 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic fragment screening-based study of a novel FAD-dependent oxidoreductase from Chaetomium thermophilum.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
6ZE3
 
 | | FAD-dependent oxidoreductase from Chaetomium thermophilum in complex with fragment (4-methoxycarbonylphenyl)methylazanium | | Descriptor: | (4-methoxycarbonylphenyl)methylazanium, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Svecova, L, Skalova, T, Kolenko, P, Koval, T, Oestergaard, L.H, Dohnalek, J. | | Deposit date: | 2020-06-16 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystallographic fragment screening-based study of a novel FAD-dependent oxidoreductase from Chaetomium thermophilum.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
6ZE2
 
 | | FAD-dependent oxidoreductase from Chaetomium thermophilum | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, ... | | Authors: | Svecova, L, Skalova, T, Kolenko, P, Koval, T, Oestergaard, L.H, Dohnalek, J. | | Deposit date: | 2020-06-16 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Crystallographic fragment screening-based study of a novel FAD-dependent oxidoreductase from Chaetomium thermophilum.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
4KC8
 
 | | Crystal Structure of Endo-1,5-alpha-L-arabinanase from Thermotoga petrophila RKU-1 in complex with TRIS | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Glycoside hydrolase, ... | | Authors: | Nascimento, A.F.Z, Polo, C.C, Santos, C.R, Costa, M.C.M.F, Mesa, A.N, Prade, R.A, Ruller, R, Squina, F.M, Murakami, M.T. | | Deposit date: | 2013-04-24 | | Release date: | 2014-02-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Mechanistic strategies for catalysis adopted by evolutionary distinct family 43 arabinanases.
J.Biol.Chem., 289, 2014
|
|
5L1O
 
 | | X-ray Structure of Cytochrome P450 PntM with Pentalenolactone F | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Pentalenolactone synthase, pentalenolactone F | | Authors: | Duan, L, Jogl, G, Cane, D.E. | | Deposit date: | 2016-07-29 | | Release date: | 2016-09-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | The Cytochrome P450-Catalyzed Oxidative Rearrangement in the Final Step of Pentalenolactone Biosynthesis: Substrate Structure Determines Mechanism.
J.Am.Chem.Soc., 138, 2016
|
|
5L1Q
 
 | | X-ray Structure of Cytochrome P450 PntM with Dihydropentalenolactone F | | Descriptor: | Dihydropentalenolactone F, PROTOPORPHYRIN IX CONTAINING FE, Pentalenolactone synthase | | Authors: | Duan, L, Jogl, G, Cane, D.E. | | Deposit date: | 2016-07-29 | | Release date: | 2016-09-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | The Cytochrome P450-Catalyzed Oxidative Rearrangement in the Final Step of Pentalenolactone Biosynthesis: Substrate Structure Determines Mechanism.
J.Am.Chem.Soc., 138, 2016
|
|
5L1S
 
 | |
5L1W
 
 | |
5L1V
 
 | |
5L1U
 
 | |
5L1R
 
 | | X-ray Structure of the Substrate-free Cytochrome P450 PntM | | Descriptor: | BICINE, PROTOPORPHYRIN IX CONTAINING FE, Pentalenolactone synthase | | Authors: | Duan, L, Jogl, G, Cane, D.E. | | Deposit date: | 2016-07-29 | | Release date: | 2016-09-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Cytochrome P450-Catalyzed Oxidative Rearrangement in the Final Step of Pentalenolactone Biosynthesis: Substrate Structure Determines Mechanism.
J.Am.Chem.Soc., 138, 2016
|
|
