6TZE
 
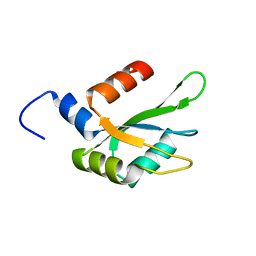 | | Human CstF-64 RRM mutant - D50A | | Descriptor: | Cleavage stimulation factor subunit 2 | | Authors: | Latham, M.P, Masoumzadeh, E. | | Deposit date: | 2019-08-12 | | Release date: | 2020-08-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A missense mutation in the CSTF2 gene that impairs the function of the RNA recognition motif and causes defects in 3' end processing is associated with intellectual disability in humans.
Nucleic Acids Res., 48, 2020
|
|
7S3A
 
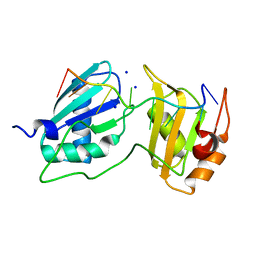 | |
7S3B
 
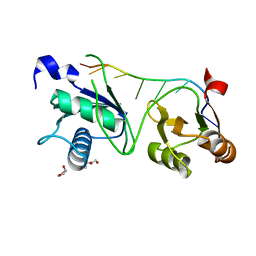 | |
7S3C
 
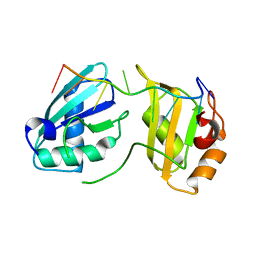 | |
5ZUH
 
 | |
1CVJ
 
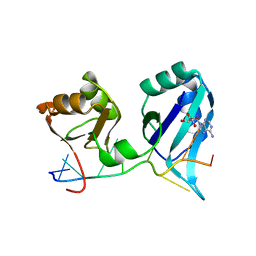 | | X-RAY CRYSTAL STRUCTURE OF THE POLY(A)-BINDING PROTEIN IN COMPLEX WITH POLYADENYLATE RNA | | Descriptor: | 5'-R(*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3', ADENOSINE MONOPHOSPHATE, POLYADENYLATE BINDING PROTEIN 1 | | Authors: | Deo, R.C, Bonanno, J.B, Sonenberg, N, Burley, S.K. | | Deposit date: | 1999-08-23 | | Release date: | 1999-10-04 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Recognition of polyadenylate RNA by the poly(A)-binding protein.
Cell(Cambridge,Mass.), 98, 1999
|
|
1D9A
 
 | | SOLUTION STRUCTURE OF THE SECOND RNA-BINDING DOMAIN (RBD2) OF HU ANTIGEN C (HUC) | | Descriptor: | HU ANTIGEN C | | Authors: | Inoue, M, Muto, Y, Sakamoto, H, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1999-10-26 | | Release date: | 2000-04-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR studies on functional structures of the AU-rich element-binding domains of Hu antigen C.
Nucleic Acids Res., 28, 2000
|
|
1D8Z
 
 | | SOLUTION STRUCTURE OF THE FIRST RNA-BINDING DOMAIN (RBD1) OF HU ANTIGEN C (HUC) | | Descriptor: | HU ANTIGEN C | | Authors: | Inoue, M, Muto, Y, Sakamoto, H, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1999-10-26 | | Release date: | 2000-04-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR studies on functional structures of the AU-rich element-binding domains of Hu antigen C.
Nucleic Acids Res., 28, 2000
|
|
1DZ5
 
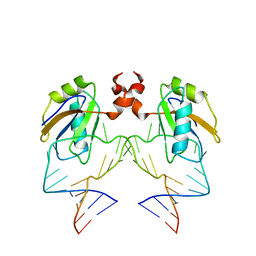 | | The NMR structure of the 38KDa U1A protein-PIE RNA complex reveals the basis of cooperativity in regulation of polyadenylation by human U1A protein | | Descriptor: | PIE, RNA (5'-R(*GP*AP*GP*AP*CP*AP*UP*UP*GP*CP*AP*CP*CP* CP*GP*GP*AP*GP*UP*CP*UP*C)-3'), U1 SMALL NUCLEAR RIBONUCLEOPROTEIN A | | Authors: | Varani, L, Gunderson, S.I, Mattaj, I.W, Kay, L.E, Neuhaus, D, Varani, G. | | Deposit date: | 2000-02-16 | | Release date: | 2000-03-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The NMR Structure of the 38kDa U1A Protein-Pie RNA Complex Reveals the Basis of Cooperativity in Regulation of Polyadenylation by Human U1A Protein
Nat.Struct.Biol., 7, 2000
|
|
1DRZ
 
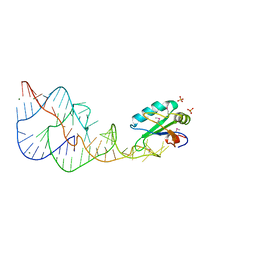 | | U1A SPLICEOSOMAL PROTEIN/HEPATITIS DELTA VIRUS GENOMIC RIBOZYME COMPLEX | | Descriptor: | MAGNESIUM ION, PROTEIN (U1 SMALL RIBONUCLEOPROTEIN A), RNA (HEPATITIS DELTA VIRUS GENOMIC RIBOZYME), ... | | Authors: | Ferre-D'Amare, A.R, Zhou, K, Doudna, J.A. | | Deposit date: | 1998-09-01 | | Release date: | 1999-02-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a hepatitis delta virus ribozyme.
Nature, 395, 1998
|
|
8RZV
 
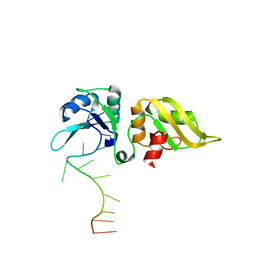 | |
7LHX
 
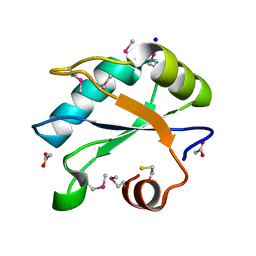 | | Human U1A protein with F37M and F77M mutations for improved phasing | | Descriptor: | ACETATE ION, BETA-MERCAPTOETHANOL, SODIUM ION, ... | | Authors: | Jenkins, J.L, Lippa, G.M, Wedekind, J.E. | | Deposit date: | 2021-01-26 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Affinity and Structural Analysis of the U1A RNA Recognition Motif with Engineered Methionines to Improve Experimental Phasing
Crystals, 11, 2021
|
|
2A3J
 
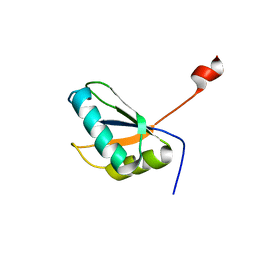 | | Structure of URNdesign, a complete computational redesign of human U1A protein | | Descriptor: | U1 small nuclear ribonucleoprotein A | | Authors: | Varani, G, Dobson, N, Dantas, G, Baker, D. | | Deposit date: | 2005-06-24 | | Release date: | 2006-06-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High-Resolution Structural Validation of the Computational Redesign of Human U1A Protein
Structure, 14, 2006
|
|
2AYM
 
 | |
2B0G
 
 | |
1ZZN
 
 | |
5SZW
 
 | | NMR solution structure of the RRM1 domain of the post-transcriptional regulator HuR | | Descriptor: | ELAV-like protein 1 | | Authors: | Lixa, C, Mujo, A, Jendiroba, K.A, Almeida, F.C.L, Lima, L.M.T.R, Pinheiro, A.S. | | Deposit date: | 2016-08-15 | | Release date: | 2017-09-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Oligomeric transition and dynamics of RNA binding by the HuR RRM1 domain in solution.
J. Biomol. NMR, 72, 2018
|
|
1L3K
 
 | | UP1, THE TWO RNA-RECOGNITION MOTIF DOMAIN OF HNRNP A1 | | Descriptor: | HETEROGENEOUS NUCLEAR RIBONUCLEOPROTEIN A1 | | Authors: | Vitali, J, Ding, J, Jiang, J, Zhang, Y, Krainer, A.R, Xu, R.-M. | | Deposit date: | 2002-02-27 | | Release date: | 2002-04-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Correlated alternative side chain conformations in the RNA-recognition motif of heterogeneous nuclear ribonucleoprotein A1.
Nucleic Acids Res., 30, 2002
|
|
1M5O
 
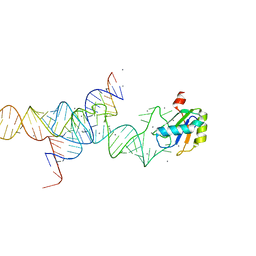 | | Transition State Stabilization by a Catalytic RNA | | Descriptor: | CALCIUM ION, RNA SUBSTRATE, RNA HAIRPIN RIBOZYME, ... | | Authors: | Rupert, P.B, Massey, A.P, Sigurdsson, S.T, Ferre-D'Amare, A.R. | | Deposit date: | 2002-07-09 | | Release date: | 2002-10-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Transition state stabilization by a catalytic RNA
Science, 298, 2002
|
|
5T9P
 
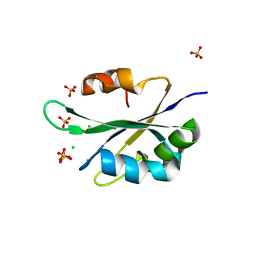 | |
1M5P
 
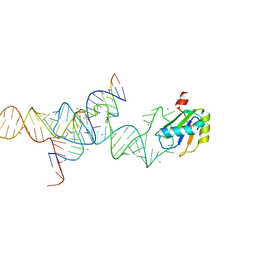 | | Transition State Stabilization by a Catalytic RNA | | Descriptor: | CALCIUM ION, RNA HAIRPIN RIBOZYME, RNA INHIBITOR SUBSTRATE, ... | | Authors: | Rupert, P.B, Massey, A, Sigurdsson, S.T, Ferre-D'Amare, A.R. | | Deposit date: | 2002-07-09 | | Release date: | 2002-10-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Transition state stabilization by a catalytic RNA
Science, 298, 2002
|
|
1JMT
 
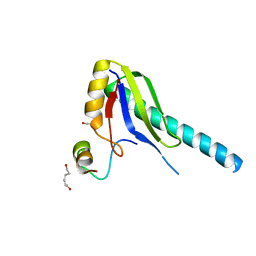 | | X-ray Structure of a Core U2AF65/U2AF35 Heterodimer | | Descriptor: | HEXANE-1,6-DIOL, SPLICING FACTOR U2AF 35 KDA SUBUNIT, SPLICING FACTOR U2AF 65 KDA SUBUNIT | | Authors: | Kielkopf, C.L, Rodionova, N.A, Green, M.R, Burley, S.K. | | Deposit date: | 2001-07-19 | | Release date: | 2001-09-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A novel peptide recognition mode revealed by the X-ray structure of a core U2AF35/U2AF65 heterodimer.
Cell(Cambridge,Mass.), 106, 2001
|
|
5TBX
 
 | | hnRNP A18 RNA Recognition Motif | | Descriptor: | ACETATE ION, Cold-inducible RNA-binding protein, NICKEL (II) ION | | Authors: | Coburn, K.M, Melville, Z, Aligholizadeh, E, Roth, B.M, Varney, K.M, Weber, D.J. | | Deposit date: | 2016-09-13 | | Release date: | 2017-04-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.767 Å) | | Cite: | Crystal structure of the human heterogeneous ribonucleoprotein A18 RNA-recognition motif.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
5TKZ
 
 | | MEC-8 N-terminal RRM bound to tandem GCAC ligand | | Descriptor: | DNA (5'-D(*AP*GP*CP*AP*CP*AP*TP*TP*TP*TP*TP*TP*TP*TP*AP*GP*CP*AP*CP*A)-3'), Mec-8 protein | | Authors: | Soufari, H, Mackereth, C.D. | | Deposit date: | 2016-10-10 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.529 Å) | | Cite: | Conserved binding of GCAC motifs by MEC-8, couch potato, and the RBPMS protein family.
RNA, 23, 2017
|
|
5UZG
 
 | | Crystal structure of Glorund qRRM1 domain | | Descriptor: | AT27789p, GLYCEROL, SULFATE ION | | Authors: | Teramoto, T, Hall, T.M.T. | | Deposit date: | 2017-02-26 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.541 Å) | | Cite: | The Drosophila hnRNP F/H Homolog Glorund Uses Two Distinct RNA-Binding Modes to Diversify Target Recognition.
Cell Rep, 19, 2017
|
|
