5TMH
 
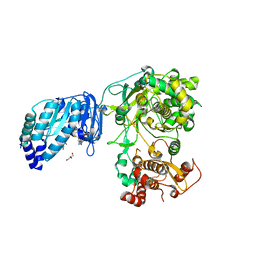 | | Structure of Zika virus NS5 | | Descriptor: | GLYCEROL, Polyprotein, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Wang, B, Tan, X, Song, J. | | Deposit date: | 2016-10-12 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.278 Å) | | Cite: | The structure of Zika virus NS5 reveals a conserved domain conformation.
Nat Commun, 8, 2017
|
|
5TTD
 
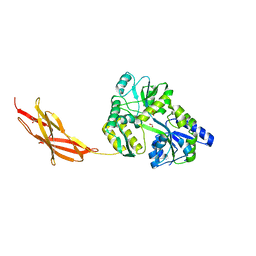 | | Minor pilin FctB from S. pyogenes with engineered intramolecular isopeptide bond | | Descriptor: | FORMIC ACID, Maltose-binding periplasmic protein,Pilin isopeptide linkage domain protein, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Young, P.G, Kwon, H, Squire, C.J, Baker, E.N. | | Deposit date: | 2016-11-02 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Engineering a Lys-Asn isopeptide bond into an immunoglobulin-like protein domain enhances its stability.
Sci Rep, 7, 2017
|
|
5TKY
 
 | | Crystal structure of the co-translational Hsp70 chaperone Ssb in the ATP-bound, open conformation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Putative uncharacterized protein | | Authors: | Gumiero, A, Gese, G.V, Weyer, F.A, Lapouge, K, Sinning, I. | | Deposit date: | 2016-10-10 | | Release date: | 2016-11-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Interaction of the cotranslational Hsp70 Ssb with ribosomal proteins and rRNA depends on its lid domain.
Nat Commun, 7, 2016
|
|
5TUJ
 
 | |
5UKD
 
 | |
4HEC
 
 | |
9EVX
 
 | | cryoEM structure of Photosystem II averaged across S2-S3 states at 1.71 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Hussein, R, Graca, A, Zouni, A, Messinger, J, Schroder, W.P. | | Deposit date: | 2024-04-02 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (1.71 Å) | | Cite: | Cryo-electron microscopy reveals hydrogen positions and water networks in photosystem II.
Science, 384, 2024
|
|
8S54
 
 | | RNA polymerase II early elongation complex bound to TFIIE and TFIIF - state b (composite structure) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit RPB11-a, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Zhan, Y, Grabber, F, Oberbeckmann, E, Dienemann, C, Cramer, P. | | Deposit date: | 2024-02-22 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Three-step mechanism of promoter escape by RNA polymerase II.
Mol.Cell, 84, 2024
|
|
8S5N
 
 | | RNA polymerase II core initially transcribing complex with an ordered RNA of 12 nt | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit RPB11-a, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Zhan, Y, Grabbe, F, Oberbeckmann, E, Dienemann, C, Cramer, P. | | Deposit date: | 2024-02-24 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Three-step mechanism of promoter escape by RNA polymerase II.
Mol.Cell, 84, 2024
|
|
8QTQ
 
 | | Thermostable WW domain | | Descriptor: | WW domain | | Authors: | Kovermann, M, Thomas, F. | | Deposit date: | 2023-10-13 | | Release date: | 2024-07-03 | | Method: | SOLUTION NMR | | Cite: | Thermostable WW-Domain Scaffold to Design Functional beta-Sheet Miniproteins.
J.Am.Chem.Soc., 2024
|
|
8S55
 
 | | RNA polymerase II early elongation complex bound to TFIIE and TFIIF - state a (composite structure) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit RPB11-a, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Zhan, Y, Grabbe, F, Oberbeckmann, E, Dienemann, C, Cramer, P. | | Deposit date: | 2024-02-22 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Three-step mechanism of promoter escape by RNA polymerase II.
Mol.Cell, 84, 2024
|
|
8S51
 
 | | RNA polymerase II core initially transcribing complex with an ordered RNA of 8 nt | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit RPB11-a, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Zhan, Y, Grabbe, F, Oberbeckmann, E, Dienemann, C, Cramer, P. | | Deposit date: | 2024-02-22 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Three-step mechanism of promoter escape by RNA polymerase II.
Mol.Cell, 84, 2024
|
|
8S52
 
 | | RNA polymerase II core initially transcribing complex with an ordered RNA of 10 nt | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit RPB11-a, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Zhan, Y, Grabbe, F, Oberbeckmann, E, Dienemann, C, Cramer, P. | | Deposit date: | 2024-02-22 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Three-step mechanism of promoter escape by RNA polymerase II.
Mol.Cell, 84, 2024
|
|
5E7C
 
 | | Macromolecular diffractive imaging using imperfect crystals - Bragg data | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Ayyer, K, Yefanov, O, Oberthuer, D, Roy-Chowdhury, S, Galli, L, Mariani, V, Basu, S, Coe, J, Conrad, C.E, Fromme, R, Schaffner, A, Doerner, K, James, D, Kupitz, C, Metz, M, Nelson, G, Xavier, P.L, Beyerlein, K.R, Schmidt, M, Sarrou, I, Spence, J.C.H, Weierstall, U, White, T.A, Yang, J.-H, Zhao, Y, Liang, M, Aquila, A, Hunter, M.S, Robinson, J.S, Koglin, J.E, Boutet, S, Fromme, P, Barty, A, Chapman, H.N. | | Deposit date: | 2015-10-12 | | Release date: | 2016-02-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Macromolecular diffractive imaging using imperfect crystals.
Nature, 530, 2016
|
|
8G3G
 
 | | CryoEM structure of yeast recombination mediator Rad52 | | Descriptor: | DNA repair and recombination protein RAD52 | | Authors: | Deveryshetty, J, Basore, K, Rau, M, Fitzpatrick, J.A.J, Antony, E. | | Deposit date: | 2023-02-07 | | Release date: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Yeast Rad52 is a homodecamer and possesses BRCA2-like bipartite Rad51 binding modes.
Nat Commun, 14, 2023
|
|
8GKC
 
 | |
8GBI
 
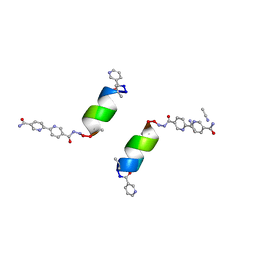 | | Porous framework formed by assembly of a bipyridyl-conjugated helical peptide | | Descriptor: | 5'-(hydrazinecarbonyl)[2,2'-bipyridine]-5-carboxamide, ACETONITRILE, LEU-(AIB)-ALA-CYS-LEU-CYS-CYS-(AIB)-LEU, ... | | Authors: | Hess, S.S, Nguyen, A.I. | | Deposit date: | 2023-02-26 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Noncovalent Peptide Assembly Enables Crystalline, Permutable, and Reactive Thiol Frameworks.
J.Am.Chem.Soc., 145, 2023
|
|
8GL0
 
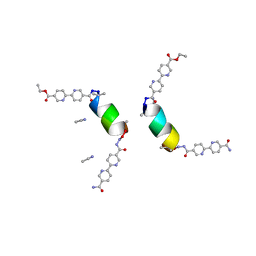 | | Porous framework formed by assembly of a bipyridyl-conjugated helical peptide | | Descriptor: | 5'-(hydrazinecarbonyl)[2,2'-bipyridine]-5-carboxamide, ACETONITRILE, LEU-AIB-ALA-CYS-LEU-CYS-GLN-AIB-LEU, ... | | Authors: | Hess, S.S, Nguyen, A.I. | | Deposit date: | 2023-03-20 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Noncovalent Peptide Assembly Enables Crystalline, Permutable, and Reactive Thiol Frameworks.
J.Am.Chem.Soc., 145, 2023
|
|
8GK1
 
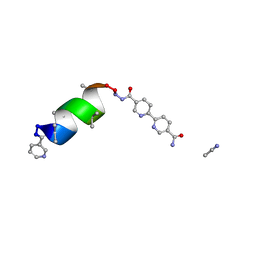 | | Porous framework formed by assembly of a bipyridyl-conjugated helical peptide | | Descriptor: | 5'-(hydrazinecarbonyl)[2,2'-bipyridine]-5-carboxamide, ACETONITRILE, LEU-AIB-ALA-AIB-LEU-AIB-GLN-AIB-LEU, ... | | Authors: | Hess, S.S, Nguyen, A.I. | | Deposit date: | 2023-03-16 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.83 Å) | | Cite: | Noncovalent Peptide Assembly Enables Crystalline, Permutable, and Reactive Thiol Frameworks.
J.Am.Chem.Soc., 145, 2023
|
|
8GKX
 
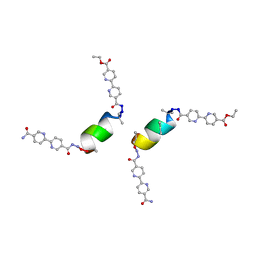 | | Porous framework formed by assembly of a bipyridyl-conjugated helical peptide | | Descriptor: | 5'-(hydrazinecarbonyl)[2,2'-bipyridine]-5-carboxamide, LEU-AIB-ALA-AIB-LEU-HIS-GLN-AIB-LEU, ethyl 5'-formyl[2,2'-bipyridine]-5-carboxylate | | Authors: | Hess, S.S, Nguyen, A.I. | | Deposit date: | 2023-03-20 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.81 Å) | | Cite: | Noncovalent Peptide Assembly Enables Crystalline, Permutable, and Reactive Thiol Frameworks.
J.Am.Chem.Soc., 145, 2023
|
|
8GJ7
 
 | | Porous framework formed by assembly of a bipyridyl-conjugated helical peptide | | Descriptor: | 5'-(hydrazinecarbonyl)[2,2'-bipyridine]-5-carboxamide, LEU-AIB-ALA-CYS-LEU-AIB-CYS-AIB-LEU, NICOTINIC ACID | | Authors: | Hess, S.S, Nguyen, A.I. | | Deposit date: | 2023-03-15 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Noncovalent Peptide Assembly Enables Crystalline, Permutable, and Reactive Thiol Frameworks.
J.Am.Chem.Soc., 145, 2023
|
|
8GBA
 
 | | Porous framework formed by assembly of a bipyridyl-conjugated helical peptide | | Descriptor: | 5'-(hydrazinecarbonyl)[2,2'-bipyridine]-5-carboxamide, LEU-(AIB)-ALA-SER-LEU-ALA-CYS-(AIB)-LEU, NICOTINIC ACID | | Authors: | Hess, S.S, Nguyen, A.I. | | Deposit date: | 2023-02-24 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Noncovalent Peptide Assembly Enables Crystalline, Permutable, and Reactive Thiol Frameworks.
J.Am.Chem.Soc., 145, 2023
|
|
8GKB
 
 | | Porous framework formed by assembly of a bipyridyl-conjugated helical peptide | | Descriptor: | 5'-(hydrazinecarbonyl)[2,2'-bipyridine]-5-carboxamide, CADMIUM ION, LEU-AIB-ALA-SER-LEU-ALA-CYS-AIB-LEU, ... | | Authors: | Hess, S.S, Nguyen, A.I. | | Deposit date: | 2023-03-17 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Noncovalent Peptide Assembly Enables Crystalline, Permutable, and Reactive Thiol Frameworks.
J.Am.Chem.Soc., 145, 2023
|
|
8GBM
 
 | | Porous framework formed by assembly of a bipyridyl-conjugated helical peptide | | Descriptor: | 5'-(hydrazinecarbonyl)[2,2'-bipyridine]-5-carboxamide, ACETATE ION, LEAD (II) ION, ... | | Authors: | Hess, S.S, Nguyen, A.I. | | Deposit date: | 2023-02-26 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Noncovalent Peptide Assembly Enables Crystalline, Permutable, and Reactive Thiol Frameworks.
J.Am.Chem.Soc., 145, 2023
|
|
8GL4
 
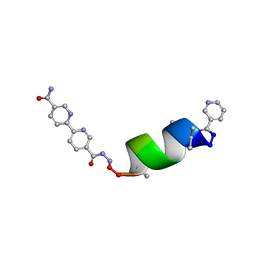 | | Porous framework formed by assembly of a bipyridyl-conjugated helical peptide | | Descriptor: | 5'-(hydrazinecarbonyl)[2,2'-bipyridine]-5-carboxamide, ACETONITRILE, LEU-AIB-ALA-CYS-LEU-CYS-GLN-AIB-LEU, ... | | Authors: | Hess, S.S, Nguyen, A.I. | | Deposit date: | 2023-03-20 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.76 Å) | | Cite: | Noncovalent Peptide Assembly Enables Crystalline, Permutable, and Reactive Thiol Frameworks.
J.Am.Chem.Soc., 145, 2023
|
|
