1HO7
 
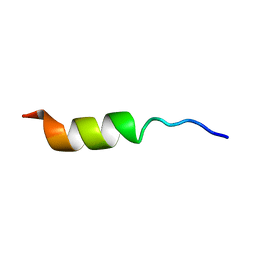 | | NMR STRUCTURE OF THE POTASSIUM CHANNEL FRAGMENT L45 IN TFE | | Descriptor: | VOLTAGE-GATED POTASSIUM CHANNEL PROTEIN | | Authors: | Ohlenschlager, O, Hojo, H, Ramachandran, R, Gorlach, M, Haris, P.I. | | Deposit date: | 2000-12-10 | | Release date: | 2002-06-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of the S4-S5 segment of the Shaker potassium channel.
Biophys.J., 82, 2002
|
|
1ALZ
 
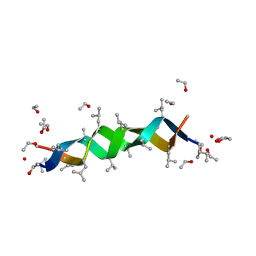 | | GRAMICIDIN D FROM BACILLUS BREVIS (ETHANOL SOLVATE) | | Descriptor: | ETHANOL, ILE-GRAMICIDIN C, VAL-GRAMICIDIN A | | Authors: | Burkhart, B.M, Pangborn, W.A, Duax, W.L, Langs, D.A. | | Deposit date: | 1997-06-06 | | Release date: | 1998-03-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.86 Å) | | Cite: | Heterodimer Formation and Crystal Nucleation of Gramicidin D
Biophys.J., 75, 1998
|
|
1AMZ
 
 | |
1HJE
 
 | |
1A4P
 
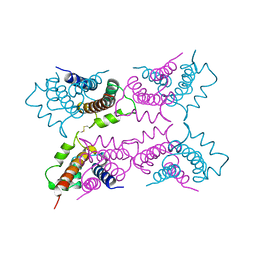 | | P11 (S100A10), LIGAND OF ANNEXIN II | | Descriptor: | S100A10 | | Authors: | Rety, S, Sopkova, J, Renouard, M, Osterloh, D, Gerke, V, Russo-Marie, F, Lewit-Bentley, A. | | Deposit date: | 1998-01-30 | | Release date: | 1998-05-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The crystal structure of a complex of p11 with the annexin II N-terminal peptide.
Nat.Struct.Biol., 6, 1999
|
|
1AL6
 
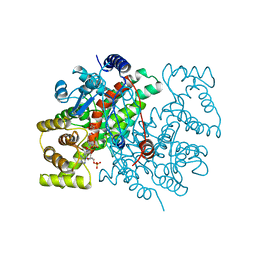 | |
1IOQ
 
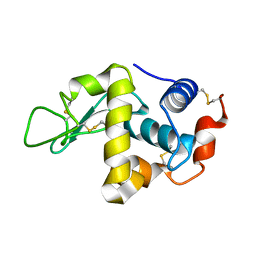 | | STABILIZATION OF HEN EGG WHITE LYSOZYME BY A CAVITY-FILLING MUTATION | | Descriptor: | LYSOZYME C | | Authors: | Ohmura, T, Ueda, T, Ootsuka, K, Saito, M, Imoto, T. | | Deposit date: | 2001-03-28 | | Release date: | 2001-04-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Stabilization of hen egg white lysozyme by a cavity-filling mutation.
Protein Sci., 10, 2001
|
|
1IOS
 
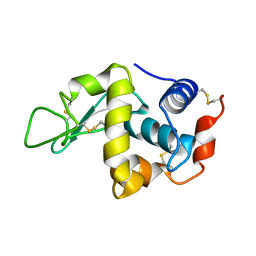 | | STABILIZATION OF HEN EGG WHITE LYSOZYME BY A CAVITY-FILLING MUTATION | | Descriptor: | LYSOZYME C | | Authors: | Ohmura, T, Ueda, T, Ootsuka, K, Saito, M, Imoto, T. | | Deposit date: | 2001-03-28 | | Release date: | 2001-04-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Stabilization of hen egg white lysozyme by a cavity-filling mutation.
Protein Sci., 10, 2001
|
|
168D
 
 | |
177D
 
 | |
1J19
 
 | | Crystal structure of the radxin FERM domain complexed with the ICAM-2 cytoplasmic peptide | | Descriptor: | 16-mer peptide from Intercellular adhesion molecule-2, radixin | | Authors: | Hamada, K, Shimizu, T, Yonemura, S, Tsukita, S, Tsukita, S, Hakoshima, T. | | Deposit date: | 2002-12-02 | | Release date: | 2003-03-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of adhesion-molecule recognition by ERM proteins revealed by the crystal structure of the radixin-ICAM-2 complex
EMBO J., 22, 2003
|
|
1IIW
 
 | |
160D
 
 | |
1IGU
 
 | |
1IJ0
 
 | |
181D
 
 | |
1ANF
 
 | | MALTODEXTRIN BINDING PROTEIN WITH BOUND MALTOSE | | Descriptor: | MALTODEXTRIN-BINDING PROTEIN, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Spurlino, J.C, Quiocho, F.A. | | Deposit date: | 1997-06-25 | | Release date: | 1997-12-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Extensive features of tight oligosaccharide binding revealed in high-resolution structures of the maltodextrin transport/chemosensory receptor.
Structure, 5, 1997
|
|
1AHO
 
 | | THE AB INITIO STRUCTURE DETERMINATION AND REFINEMENT OF A SCORPION PROTEIN TOXIN | | Descriptor: | TOXIN II | | Authors: | Smith, G.D, Blessing, R.H, Ealick, S.E, Fontecilla-Camps, J.C, Hauptman, H.A, Housset, D, Langs, D.A, Miller, R. | | Deposit date: | 1997-04-08 | | Release date: | 1997-10-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | Ab initio structure determination and refinement of a scorpion protein toxin.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
152D
 
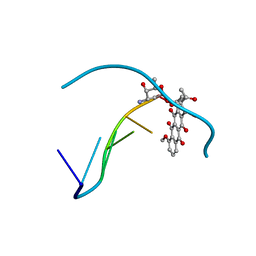 | | DIVERSITY OF WATER RING SIZE AT DNA INTERFACES: HYDRATION AND DYNAMICS OF DNA-ANTHRACYCLINE COMPLEXES | | Descriptor: | DAUNOMYCIN, DNA (5'-D(*CP*GP*AP*TP*CP*G)-3') | | Authors: | Lipscomb, L.A, Peek, M.E, Zhou, F.X, Bertrand, J.A, VanDerveer, D, Williams, L.D. | | Deposit date: | 1993-12-13 | | Release date: | 1994-05-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Water ring structure at DNA interfaces: hydration and dynamics of DNA-anthracycline complexes.
Biochemistry, 33, 1994
|
|
180D
 
 | |
187D
 
 | |
1J2U
 
 | | Creatininase Zn | | Descriptor: | SULFATE ION, ZINC ION, creatinine amidohydrolase | | Authors: | Yoshimoto, T, Tanaka, N, Kanada, N, Inoue, T, Nakajima, Y, Haratake, M, Nakamura, K.T, Xu, Y, Ito, K. | | Deposit date: | 2003-01-11 | | Release date: | 2004-01-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of creatininase reveal the substrate binding site and provide an insight into the catalytic mechanism
J.Mol.Biol., 337, 2004
|
|
153D
 
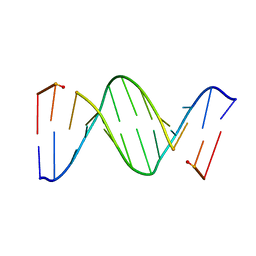 | | CRYSTAL STRUCTURE OF A MISPAIRED DODECAMER, D(CGAGAATTC(O6ME)GCG)2, CONTAINING A CARCINOGENIC O6-METHYLGUANINE | | Descriptor: | DNA (5'-D(*CP*GP*AP*GP*AP*AP*TP*TP*CP*(6OG)P*CP*G)-3') | | Authors: | Ginell, S.L, Vojtechovsky, J, Gaffney, B, Jones, R, Berman, H.M. | | Deposit date: | 1993-12-16 | | Release date: | 1994-01-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of a mispaired dodecamer, d(CGAGAATTC(O6Me)GCG)2, containing a carcinogenic O6-methylguanine
Biochemistry, 33, 1994
|
|
1HSO
 
 | | HUMAN ALPHA ALCOHOL DEHYDROGENASE (ADH1A) | | Descriptor: | 4-IODOPYRAZOLE, CLASS I ALCOHOL DEHYDROGENASE 1, ALPHA SUBUNIT, ... | | Authors: | Niederhut, M.S, Gibbons, B.J, Perez-Miller, S, Hurley, T.D. | | Deposit date: | 2000-12-27 | | Release date: | 2001-01-10 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three-dimensional structures of the three human class I alcohol dehydrogenases.
Protein Sci., 10, 2001
|
|
1HRU
 
 | | THE STRUCTURE OF THE YRDC GENE PRODUCT FROM E.COLI | | Descriptor: | PHOSPHATE ION, YRDC GENE PRODUCT | | Authors: | Teplova, M, Tereshko, V, Sanishvili, R, Joachimiak, A, Bushueva, T, Anderson, W.F, Egli, M, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2000-12-21 | | Release date: | 2001-01-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of the yrdC gene product from Escherichia coli reveals a new fold and suggests a role in RNA binding.
Protein Sci., 9, 2000
|
|
