8OE2
 
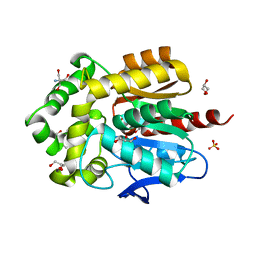 | | Structure of hyperstable haloalkane dehalogenase variant DhaA223 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Marek, M. | | Deposit date: | 2023-03-10 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Advancing Enzyme's Stability and Catalytic Efficiency through Synergy of Force-Field Calculations, Evolutionary Analysis, and Machine Learning.
Acs Catalysis, 13, 2023
|
|
4NG9
 
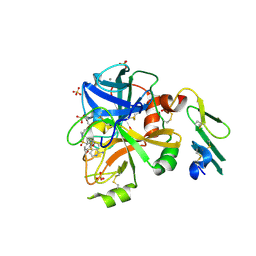 | |
1UUB
 
 | |
7NFR
 
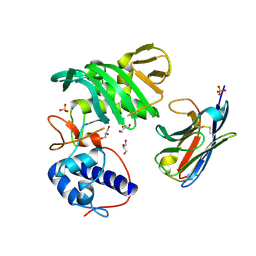 | | Fujian capmidlink domain in complex with Nb8194 | | Descriptor: | GLYCEROL, NB8194, Polymerase basic protein 2, ... | | Authors: | Keown, J.R, Grimes, J.M, Fodor, E. | | Deposit date: | 2021-02-07 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Mapping inhibitory sites on the RNA polymerase of the 1918 pandemic influenza virus using nanobodies.
Nat Commun, 13, 2022
|
|
7NFQ
 
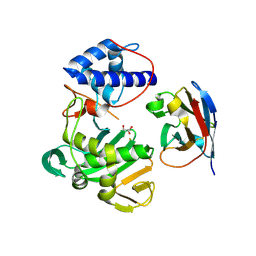 | | Fujian capmidlink domain in complex with Nb8193 | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Nb8193, ... | | Authors: | Keown, J.R, Grimes, J.M, Fodor, E. | | Deposit date: | 2021-02-07 | | Release date: | 2021-12-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Mapping inhibitory sites on the RNA polymerase of the 1918 pandemic influenza virus using nanobodies.
Nat Commun, 13, 2022
|
|
5AL2
 
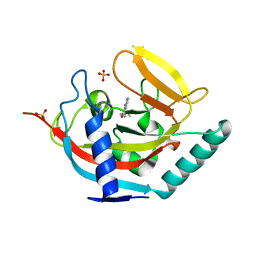 | | Crystal structure of TNKS2 in complex with 2-(4-(propan-2-yl)phenyl)- 1H,2H,3H,4H-pyrido(2,3-d)pyrimidin-4-one | | Descriptor: | (2S)-2-(4-propan-2-ylphenyl)-2,3-dihydro-1H-pyrido[2,3-d]pyrimidin-4-one, GLYCEROL, SULFATE ION, ... | | Authors: | Nkizinkiko, Y, Lehtio, L. | | Deposit date: | 2015-03-06 | | Release date: | 2015-07-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of Potent and Selective Nonplanar Tankyrase Inhibiting Nicotinamide Mimics.
Bioorg.Med.Chem., 23, 2015
|
|
1UUA
 
 | |
2REO
 
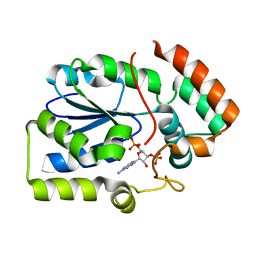 | | Crystal structure of human sulfotransferase 1C3 (Sult1C3) in complex with PAP | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, Putative sulfotransferase 1C3 | | Authors: | Tempel, W, Pan, P, Dong, A, Loppnau, P, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-09-26 | | Release date: | 2007-10-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.651 Å) | | Cite: | Crystal structure of human sulfotransferase 1C3 (Sult1C3) in complex with PAP.
To be Published
|
|
7SG5
 
 | | Structure of PfCSP peptide 21 with antibody CIS43_Var2 | | Descriptor: | CIS43_Var2 Fab Heavy chain, CIS43_Var2 Fab Light chain, GLYCEROL, ... | | Authors: | Tripathi, P, Kwong, P.D. | | Deposit date: | 2021-10-05 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Highly protective antimalarial antibodies via precision library generation and yeast display screening.
J.Exp.Med., 219, 2022
|
|
7YOP
 
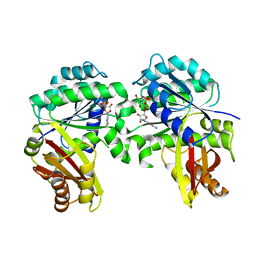 | | Spiroplasma melliferum FtsZ bound to GMPPNP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Chakraborty, J, Pananghat, G. | | Deposit date: | 2022-08-01 | | Release date: | 2023-08-16 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.20003128 Å) | | Cite: | Dynamics of interdomain rotation facilitates FtsZ filament assembly.
J.Biol.Chem., 300, 2024
|
|
7YSZ
 
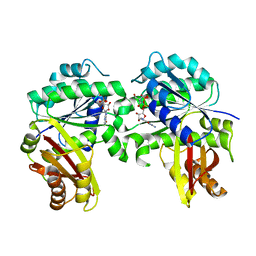 | | Spiroplasma melliferum FtsZ bound to GDP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Chakraborty, J, Pananghat, G. | | Deposit date: | 2022-08-13 | | Release date: | 2023-08-16 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.3000412 Å) | | Cite: | Dynamics of interdomain rotation facilitates FtsZ filament assembly.
J.Biol.Chem., 300, 2024
|
|
1ZG8
 
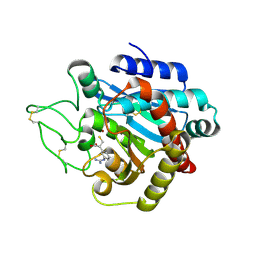 | | Crystal Structure of (R)-2-(3-{[amino(imino)methyl]amino}phenyl)-3-sulfanylpropanoic acid Bound to Activated Porcine Pancreatic Carboxypeptidase B | | Descriptor: | (2R)-2-(3-{[AMINO(IMINO)METHYL]AMINO}PHENYL)-3-SULFANYLPROPANOIC ACID, ZINC ION, procarboxypeptidase B | | Authors: | Adler, M, Bryant, J, Buckman, B, Islam, I, Larsen, B, Finster, S, Kent, L, May, K, Mohan, R, Yuan, S, Whitlow, M. | | Deposit date: | 2005-04-20 | | Release date: | 2005-07-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of potent thiol-based inhibitors bound to carboxypeptidase b.
Biochemistry, 44, 2005
|
|
4K2Z
 
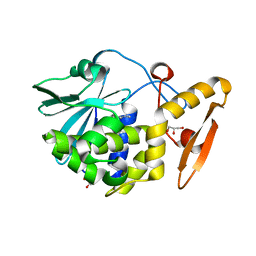 | | Crystal structure of the complex of type I Ribosome inactivating protein from Momordica balsamina with Methylethylamine at 1.80 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, METHYLETHYLAMINE, ... | | Authors: | Yamini, S, Pandey, S, Kushwaha, G.S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-04-10 | | Release date: | 2013-05-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the complex of type I Ribosome inactivating protein from Momordica balsamina with Methylethylamine at 1.80 A resolution
To be Published
|
|
4KMK
 
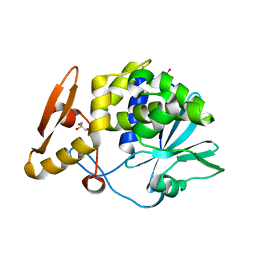 | | Crystal structure of Ribosome Inactivating protein from Momordica balsamina at 1.65 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, rRNA N-glycosidase | | Authors: | Yamini, S, Pandey, S, Kushwaha, G.S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-05-08 | | Release date: | 2013-05-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of Ribosome Inactivating protein from Momordica balsamina at 1.65 A resolution
To be Published
|
|
7AT6
 
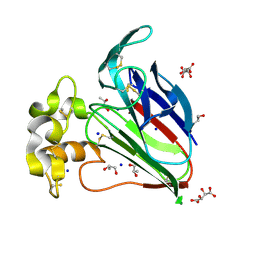 | | Structure of thaumatin collected by femtosecond serial crystallography on a COC membrane | | Descriptor: | L(+)-TARTARIC ACID, R-1,2-PROPANEDIOL, SODIUM ION, ... | | Authors: | Martiel, I, Marsh, M, Vera, L, Huang, C.Y, Olieric, V, Leonarski, P, Nass, K, Padeste, C, Karpik, A, Wang, M, Pedrini, B. | | Deposit date: | 2020-10-29 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Commissioning results from the SwissMX instrument for fixed target macromolecular crystallography at SwissFEL
To Be Published
|
|
6LFQ
 
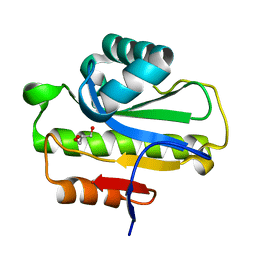 | | Crystal structure of Poa1p in apo form | | Descriptor: | ADP-ribose 1''-phosphate phosphatase, GLYCEROL | | Authors: | Chiu, Y.C, Hsu, C.H. | | Deposit date: | 2019-12-03 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.359 Å) | | Cite: | Expanding the Substrate Specificity of Macro Domains toward 3''-Isomer of O-Acetyl-ADP-ribose
Acs Catalysis, 11, 2021
|
|
4XXS
 
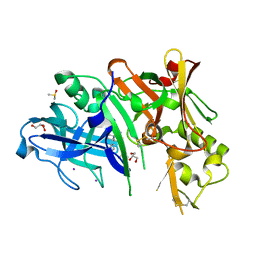 | | Crystal structure of BACE1 with a pyrazole-substituted tetrahydropyran thioamidine | | Descriptor: | (4aR,6R,8aS)-8a-(2,4-difluorophenyl)-6-(1-methyl-1H-pyrazol-4-yl)-4,4a,5,6,8,8a-hexahydropyrano[3,4-d][1,3]thiazin-2-amine, Beta-secretase 1, DIMETHYL SULFOXIDE, ... | | Authors: | Parris, K.D, Pandit, J. | | Deposit date: | 2015-01-30 | | Release date: | 2015-04-01 | | Last modified: | 2015-04-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Utilizing Structures of CYP2D6 and BACE1 Complexes To Reduce Risk of Drug-Drug Interactions with a Novel Series of Centrally Efficacious BACE1 Inhibitors.
J.Med.Chem., 58, 2015
|
|
6S22
 
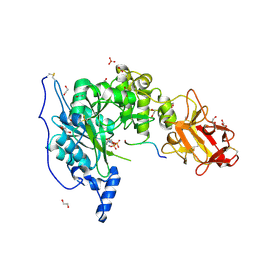 | | Crystal structure of the TgGalNAc-T3 in complex with UDP, manganese and FGF23c | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | de las Rivas, M, Daniel, E.J.P, Narimatsu, Y, Companon, I, Kato, K, Hermosilla, P, Thureau, A, Ceballos-Laita, L, Coelho, H, Bernado, P, Marcelo, F, Hansen, L, Lostao, A, Corzana, F, Clausen, H, Gerken, T.A, Hurtado-Guerrero, R. | | Deposit date: | 2019-06-20 | | Release date: | 2019-12-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Molecular basis for fibroblast growth factor 23 O-glycosylation by GalNAc-T3.
Nat.Chem.Biol., 16, 2020
|
|
2WDO
 
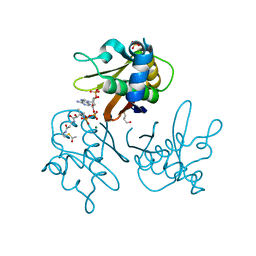 | | Crystal structure of the S. coelicolor AcpS in complex with acetyl- CoA at 1.5 A | | Descriptor: | ACETYL COENZYME *A, COENZYME A, GLYCEROL, ... | | Authors: | Dall'Aglio, P, Arthur, C, Crump, M.P, Crosby, J, Hadfield, A.T. | | Deposit date: | 2009-03-25 | | Release date: | 2010-04-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Analysis of Streptomyces Coelicolor Phosphopantetheinyl Transferase, Acps, Reveals the Basis for Relaxed Substrate Specificity.
Biochemistry, 50, 2011
|
|
3GZX
 
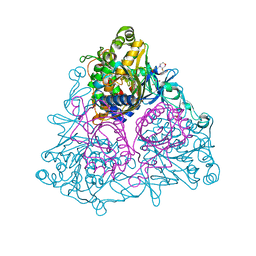 | | Crystal Structure of the Biphenyl Dioxygenase in complex with Biphenyl from Comamonas testosteroni Sp. Strain B-356 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BIPHENYL, Biphenyl dioxygenase subunit alpha, ... | | Authors: | Kumar, P, Colbert, C.L, Bolin, J.T. | | Deposit date: | 2009-04-08 | | Release date: | 2010-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural Characterization of Pandoraea pnomenusa B-356 Biphenyl Dioxygenase Reveals Features of Potent Polychlorinated Biphenyl-Degrading Enzymes
Plos One, 8, 2013
|
|
5Z5D
 
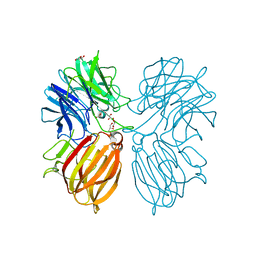 | | Crystal structure of a thermostable glycoside hydrolase family 43 {beta}-1,4-xylosidase from Geobacillus thermoleovorans IT-08 | | Descriptor: | Beta-xylosidase, CALCIUM ION, GLYCEROL | | Authors: | Rohman, A, van Oosterwijk, N, Puspaningsih, N.N.T, Dijkstra, B.W. | | Deposit date: | 2018-01-17 | | Release date: | 2018-04-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of product inhibition by arabinose and xylose of the thermostable GH43 beta-1,4-xylosidase from Geobacillus thermoleovorans IT-08.
PLoS ONE, 13, 2018
|
|
4N9V
 
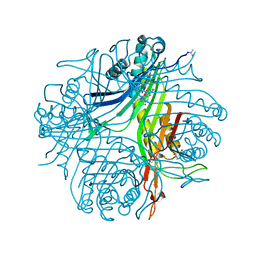 | | High resolution x-ray structure of urate oxidase in complex with 8-azaxanthine | | Descriptor: | 8-AZAXANTHINE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Oksanen, E, Blakeley, M.P, Budayova-Spano, M. | | Deposit date: | 2013-10-21 | | Release date: | 2014-02-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The neutron structure of urate oxidase resolves a long-standing mechanistic conundrum and reveals unexpected changes in protonation.
Plos One, 9, 2014
|
|
4OYR
 
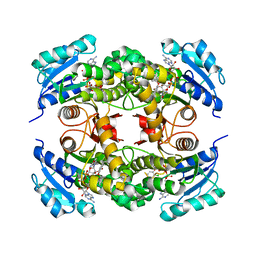 | | Competition of the small inhibitor PT91 with large fatty acyl substrate of the Mycobacterium tuberculosis enoyl-ACP reductase InhA by induced substrate-binding loop refolding | | Descriptor: | 2-(2-chloranylphenoxy)-5-hexyl-phenol, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Li, H.J, Pan, P, Lai, C.T, Liu, N, Garcia-Diaz, M, Simmerling, C, Tonge, P.J. | | Deposit date: | 2014-02-13 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2995 Å) | | Cite: | A Structural and Energetic Model for the Slow-Onset Inhibition of the Mycobacterium tuberculosis Enoyl-ACP Reductase InhA.
Acs Chem.Biol., 9, 2014
|
|
4K42
 
 | | Crystal structure of actin in complex with synthetic AplC tail analogue SF01 [(3R,4S,5R,6S,10R,11R,12R)-11-(acetyloxy)-1-(benzyloxy)-14-[formyl(methyl)amino]-5-hydroxy-4,6,10,12-tetramethyl-9-oxotetradecan-3-yl propanoate] | | Descriptor: | (3R,4S,5R,6S,10R,11R,12R)-11-(acetyloxy)-1-(benzyloxy)-14-[formyl(methyl)amino]-5-hydroxy-4,6,10,12-tetramethyl-9-oxotetradecan-3-yl propanoate, ADENOSINE-5'-DIPHOSPHATE, Actin, ... | | Authors: | Pereira, J.H, Petchprayoon, C, Moriarty, N.W, Fink, S.J, Cecere, G, Paterson, I, Adams, P.D, Marriott, G. | | Deposit date: | 2013-04-11 | | Release date: | 2014-10-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and biochemical studies of actin in complex with synthetic macrolide tail analogues.
Chemmedchem, 9, 2014
|
|
4OXY
 
 | | Substrate-binding loop movement with inhibitor PT10 in the tetrameric Mycobacterium tuberculosis enoyl-ACP reductase InhA | | Descriptor: | 5-hexyl-2-(2-nitrophenoxy)phenol, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Li, H.J, Sullivan, T.J, Pan, P, Lai, C.T, Liu, N, Garcia-Diaz, M, Simmerling, C, Tonge, P.J. | | Deposit date: | 2014-02-09 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3501 Å) | | Cite: | A Structural and Energetic Model for the Slow-Onset Inhibition of the Mycobacterium tuberculosis Enoyl-ACP Reductase InhA.
Acs Chem.Biol., 9, 2014
|
|
